2
A Trans-Kingdom Perspective on Animal Models and Microbiome Research
Herbert Virgin, the Edward Mallinckrodt Professor and chair of the pathology and immunology department at the Washington University School of Medicine in St. Louis, began his presentation by explicitly defining the term microbiome as the collection of all the organisms in or on a host, including viruses, bacteria, Archaea, fungi, and protists. All of these organisms interact with each other and the host in a variety of complex and meaningful ways.
The virome—Virgin’s area of academic expertise—is a permanent and dynamic contributor to the human metagenome. Every human hosts a number of active, living viruses at any given moment (collectively called “the virome”), most of which are novel and uncharacterized while perpetually shedding virus particles with few overt health consequences (Virgin, 2014).
Every human has a unique virome, whose components constantly interact with each other, with other organisms in the microbiome, and with the host, and influence both the host genotype and phenotype. Virgin hypothesized that these viruses may even be defining how our immune systems react to challenge—“if we’re chronically infected but apparently healthy, that may define our immunophenotype.” In fact, he and his colleagues have shown that mice with a latent herpesvirus infection are protected from challenge with a very high dose of Listeria monocytogenes due to increased expression of gamma interferon (IFN-γ). The same protection is observed in genetically modified mice that lack the HOIL-1 gene, which produces a severely immunocompromised phenotype that is also seen in humans (Boisson et al., 2012; MacDuff et al., 2015).
THE IMPORTANCE OF TRANS-KINGDOM INTERACTIONS
The complex nature of the interactions between microbiome and host—Virgin calls them trans-kingdom interactions—has important implications for understanding the relationship between disease genotype and phenotype. Virgin related our current understanding of the microbiome to that of the solar system, which only made sense when scientists understood the central position of the
sun rather than the Earth. Relatedly, Virgin argues, researchers now view the microbiome almost exclusively in relation to the host, when the interactions among the components of the microbiome are just as critical.
As an example of this trans-kingdom complexity, Virgin returned to the herpesvirus experiment to note that infection of these mice with a helminth induces interleukin 4 (IL-4), which in turn triggers an active viral infection, that is, reversal of the herpesvirus latency (Reese et al., 2014). The virus contains separate promoters that cause a different response to each of the two cytokines (IFN-γ and IL-4); that is, “this is not about the host controlling the virus. This is about the virus evolving promoters to leverage what the host is doing,” noted Virgin.
In a second example of a trans-kingdom interaction, Virgin discussed a series of experiments showing that bacteria can control chronic norovirus infections. Some members of the norovirus family cause severe gastrointestinal distress, while others produce an asymptomatic persistent infection of the spleen, lymph nodes, and other tissues (Nice et al., 2013). Virgin and his collaborators have shown that chronic norovirus infection in a mouse model can increase susceptibility to inflammatory bowel disease (IBD) (Cadwell et al., 2008, 2010). “You can prevent the virus-triggered pathology by giving antibiotics, indicating that the virus is interacting with bacteria,” said Virgin, noting that pre-treating the animals with antibiotics protected them from persistent infection, indicating the reliance of the virus on the bacteria to ensure persistence (Baldridge et al., 2015). Subsequent experiments showed that introduction of interferon lambda (IFN-λ) produced the same results against norovirus as exposing the mice to antibiotics (Nice et al., 2015). These were surprising findings, said Virgin, due to the implication that the innate immune system can clear an infection, which goes against the dogma that the innate immune system simply holds an infection in check until the adaptive immune system responds. He noted that other investigators have demonstrated a similar phenomenon with rotaviruses (Uchiyama et al., 2014; Zhang et al., 2014).
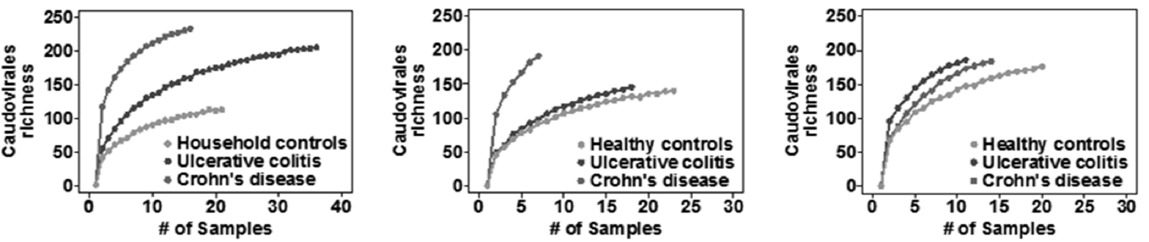
Researchers are uncertain whether such trans-kingdom interactions occur in humans, but Virgin presented evidence suggesting that they do. Examination of the bacterial microbiomes and viromes of three patient cohorts with IBD showed decreased microbiome complexity but increased virome complexibity in the IBD patients versus healthy controls due to elevated numbers of Caudiovirales (Norman et al., 2015) (see Figure 2-1). Further analysis showed that the viromes associated with Crohn’s disease differed from the viromes found in ulcerative colitis patients (both diseases are classified as IBD). Based on these results, Virgin and his colleagues have hypothesized that these diseases develop when viruses are killing beneficial bacteria, rather than in the absence of beneficial or the presence of pathogenic bacteria.
Virgin described another set of experiments which demonstrated that sequential infection of mice with a variety of viruses and one worm altered the animals’ response to yellow fever vaccination (Reese et al., 2016). These experiments, designed to mimic the early exposure to infections in humans and the subsequent changes to the immune system’s phenotype, showed that early exposure in mice versus humans causes very different phenotypes. This is an important observation regarding animal models because it could explain why mouse models do not predict human vaccine response: “Maybe mice are very representative, but we have cleaned them up to the point where we have made them nonresponsive,” said Virgin.
Virgin re-emphasized that the components of the microbiome—viruses, bacteria, worms, and other organisms—do not act in isolation. Their trans-kingdom interactions have substantial physiological effects on the host. In order to elucidate the effect of these interactions on health and disease, the field needs to conduct many more carefully controlled clinical and animal studies. In doing so, Virgin called for researchers to fully and transparently report the experimental conditions and methods used. He further highlighted the need for massive improvements in the scope and annotation quality of genome databases and called for the creation of kingdom-specific bioinformatics tools. In his opinion, the field must evolve from an associational science to one that manipulates the different -omes and tests mechanistic hypotheses, and to do so it needs to improve the quality and reproducibility of sequencing.
Concluding his presentation, Virgin called for researchers to train more students and break down the silos that keep virologists, bacteriologists, mycologists, and others from talking to one another. Integrating disciplines is essential for developing a more complete understanding of the role that the microbiome plays in health and disease.




