5
Generating, Integrating, and Accessing Digital Data
Throughout most of their history, biological collections and the physical specimens they contained were explicitly linked to the physical locations where they were housed. These biological collections consisted of specimens and their accompanying data in written records, and to access the collections users had to travel to the collection or receive specimens through the mail. That is changing now, however, as increasing numbers of biological collections have been digitized. This digitization1 of specimen data, combined with the cyberinfrastructure2 that underlies how digital data are stored, managed, and used, has fundamentally transformed the biological collections community (Ball-Damerow et al., 2019; Hedrick et al., 2020) and the work of researchers who rely on biological collections as digitization makes possible the remote examination of biological collections and greatly enhances their discoverability and usefulness.
A key component of digitization has been the development of collection databases that provide digital specimen data to aggregated data repositories, producing a global biodiversity infrastructure. Online data repositories democratize access to digital specimen data, making possible new avenues of scientific inquiry, promoting the multiplication and expansion of research collaborations and community networks, and providing a greater range of educational and training opportunities (Lacey et al., 2017; Monfils et al., 2017). A robust cyberinfrastructure can also facilitate evaluation and the development of metrics for assessing the diversity of biological collections and their impact on research and education (Meehan et al., 2019) (see Chapters 2 and 3).
Biological collections have driven increasingly integrative and collaborative science—with the potential to address a wide variety of problems from disease, such as coronavirus disease 2019 (Cook et al., 2020), to species responses to climate change (Meineke et al., 2018a)—which in turn has intensified the need for greater access to high-quality digital data. Over the past decade, a wide range of advances in the process of generating digital data of all kinds and building the cyberinfrastructure for biological collections has emerged. However, the robust cyberinfrastructure that the biological collections community requires has yet to be fully realized. This chapter focuses on the challenges of and strategies for
___________________
1 The conversion of textual, image, or sound-based specimen information to digital formats.
2 Cyberinfrastructure, a term first used by the National Science Foundation, encompasses the computing systems, repositories, advanced instruments, software, high-performance networks, and people that enable and support data acquisition, storage, management, integration, mining, analysis, visualization, and distribution (adapted from Stewart et al., 2019). See https://scholarworks.iu.edu/dspace/handle/2022/12967.
advancing the accessibility and integration of digital biological collections for research and education.
CURRENT STATE OF DIGITIZATION, DATA, AND CYBERINFRASTRUCTURE
Digitization: An Evolving Process
Biological collections encompass a diverse array of specimen data that span biological, physiological, temporal, and spatial features of the specimens. Digitization is the process of converting these analog or printed specimen data from specimen labels, field notes, card catalogs, ledgers, genetic sequences, images, audio, and video recordings, and more into digital representations. Digitization helps preserve the long-term integrity of specimens by allowing researchers to inspect metadata and digital images without having to access and physically handle the specimens while opening new avenues of data-driven research (e.g., ecological niche modeling). The biological collections community has spent decades digitizing specimen data to increase their visibility and accessibility to researchers, educators, and the general public. In fact, the digitization of specimens and associated materials and the uploading of these digital data into online platforms has long been a requirement for funding programs such as the National Science Foundation (NSF) Living Stock Collections for Biological Research program and its successor, the Collections in Support of Biological Research program, among others. In 2010 the National Evolutionary Synthesis Center workshop outlined a vision and strategic plan for a Network Integrated Biocollections Alliance to “document the nation’s biodiversity resources and create a dynamic electronic resource that will serve the country’s needs in answering critical questions” (NESCent, 2010, p. 2). At that point, it was estimated that only approximately 10 percent of all specimens in natural history collections worldwide had been digitized (Page et al., 2015). NSF responded to elements of the NIBA plan by establishing the Advancing Digitization of Biodiversity Collections (ADBC)3 program, which funds digitization efforts that coalesce around scientific questions or themes through extensive collaborative networks, called thematic collections networks (TCNs), overseen by the national coordinating center for these efforts, Integrated Digitized Biocollections (iDigBio).4 iDigBio now hosts more than 121 million digital specimen records, the majority of which were largely unavailable to users 10 years ago. Based on iDigBio’s digitized holdings compared with estimates of specimens held in U.S. collections, it is now estimated that about 30 percent of all natural history specimens in the United States have been digitized. However, there is still a long way to go until all collections have been digitized, particularly given the challenges posed by certain types of collections and the need for a workforce with both curatorial and data management skills. However, thanks to recent efforts, research using
___________________
3 See https://www.nsf.gov/funding/pgm_summ.jsp?pims_id=503559.
4 See https://www.idigbio.org.
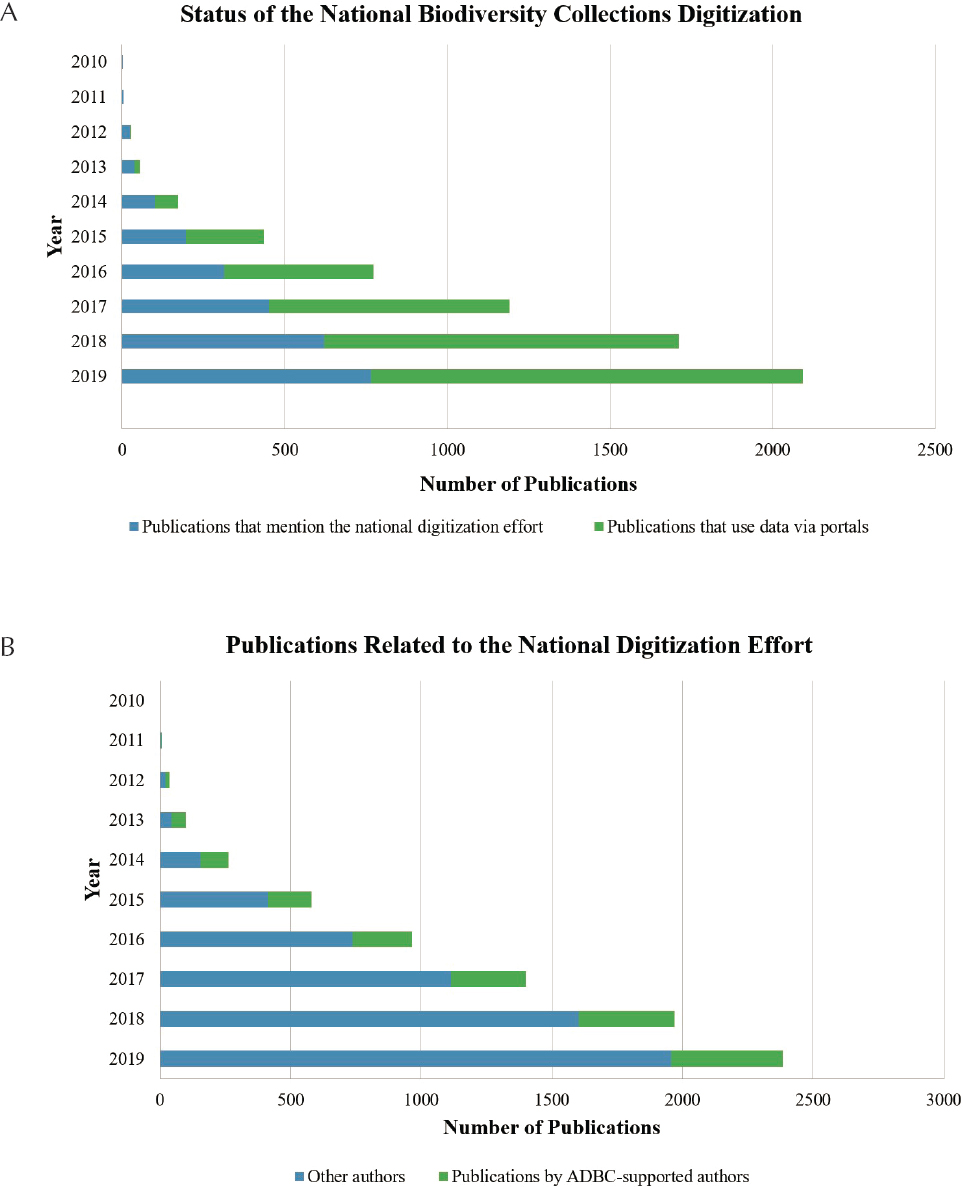
natural history collections data, as measured by citations in publications, has increased dramatically over the past decade, reflecting the increasing number of digitized collections (e.g., Ball-Damerow et al., 2019; Heberling et al., 2019) (see Figure 5-1).
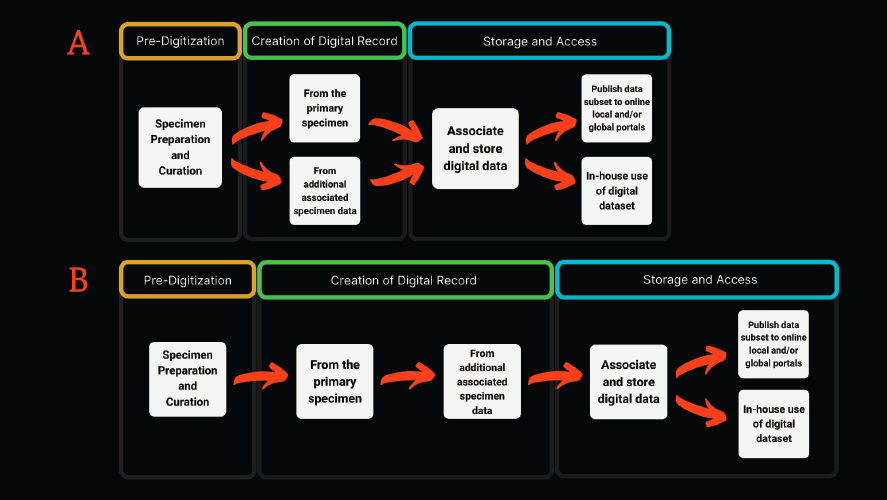
The development of digitization workflows (e.g., Haston et al., 2012; Karim et al., 2016; Nelson et al., 2012, 2015; Tulig et al., 2012) over the past decade, coupled with an emerging community of practice among collections professionals, provides a roadmap for accelerating the pace of digitization in the coming decade if sufficient funding can be made available. These digitization workflows provide institutions that house biological collections with guiding principles that can be adapted to their varied needs, collection sizes, and capabilities. Additionally, workshops organized and sponsored by iDigBio5 and others have made digitization more widely adopted, better understood, and more efficient across the natural history collections community. Living and natural history collections follow the same general digitization workflow (see Figure 5-2), with all collections providing data on the source of the specimen, date of sampling, the collector, and other attributes of provenance. However, the workflows will differ between collections due to the unique digitization priorities of each collection and the varying needs of their respective research and end-user communities.
Rapid technological advances in digitization and cyberinfrastructure have allowed a large amount of historical data to be converted into digital representations over the past 20 years. The current digitization process for existing specimens typically involves hand-entering primary data from a specimen label, field notes, card catalog, or ledger into a database, which can be time-consuming. As described later in this chapter, numerous attempts have been made to speed up this process while preserving the quality of the digital data produced. The pace of digitizing newly acquired specimens, on the other hand, is much more rapid. Specimen data are increasingly “born” digital—directly produced in digital format (e.g., GPS locations, digital spreadsheets, nucleic acid sequencing, three-dimensional images, computer tomography, etc.), which drastically reduces the amount of time required to create specimen records and integrate and share them online.
Toward Accessible and Integrated Data
Digital data from biological collections can be organized into one or more datasets that are collectively stored in local databases. At the local level, digitized collections can be easier to manage than non-digitized collections and may improve the ability of the collection managers to provide access, respond to requests, physically manage space, and allocate budget resources. Digital collection databases can be published and then accessed through online thematic, taxonomic, or geographic data portals, aggregators, and catalogs. Often, the biological collections community uses portals and aggregators interchangeably. In this report a portal is defined as the online platform that allows users to perform advanced searches on the published collections found therein. This could be a local portal to an individual collection or a portal of aggregated collections. An aggregator is the cyberinfrastructure that gathers and compiles data from published collections and makes them searchable through portals. A catalog is
___________________
5 See https://www.idigbio.org/content/workflow-modules-and-task-lists.
similar to a portal but is a term mostly used by the living collection community. Catalogs enable users to search, request, or buy specimens and materials, facilitate the collection of fees, and provide information on shipping permits, compliance with regulations, and user registration unique to living collections.
A major global portal for natural history collections is hosted by the Global Biodiversity Information Facility (GBIF), while iDigBio hosts a portal for collections primarily based in the United States, and the Atlas of Living Australia (ALA) and the Distributed System of Scientific Collections provide portals to Australian and European collections, respectively. There are also project-based portals (e.g., TCN portals, such as the SouthEast Regional Network of Expertise and Collections) and taxonomic portals (e.g., Vertnet, Fishnet, EntoWeb, iDigPaleo). The data that are available via major portals are based on common standards (e.g., Darwin Core Standards6 or the Access to Biological Collections Data7 schema). These standards help data providers share specimen data using a common terminology of fields, controlled vocabularies, and data classes that describe the taxonomic identity, collecting event, locality, collectors, geological context, and specimen attributes as well as various kinds of media (Wieczorek et al., 2012). The use of these common standards facilitates the computerized aggregation of data from multiple types of collections and the integration of specimen data with other sources of information. Users are able to search data, download results for further analysis, and integrate the downloaded data with other resources, such as environmental data (e.g., temperature, precipitation, etc.). New standards to allow for the incorporation of additional properties, called extensions, are continually being developed by the global community.
Living stock collections serve as specimen repositories and data providers for members of the research community, who interface with these collections through online databases, catalogs, and aggregators. One such centralized aggregator, the Global Catalogue of Microorganisms (GCM),8 hosted by the World Federation for Culture Collections and managed by the Chinese Academy of Sciences, facilitates the access and sharing of microbial living stock collections along with their associated data. Online platforms provide information such as available strains, genes and alleles, and genome sequences with functional annotation for the acquisition of research material. Standardized abbreviations for genes, alleles, and depositors and coordinated genome sequencing and annotation projects help make these data useful to the user community (Jarret and McCluskey, 2019). In the GCM, users can locate desired strains along with the associated metadata (e.g., date of isolation, geographic origin, growth conditions, and medium, etc.). Users can add strains to a shopping cart if they wish to acquire them for research, and by putting a strain in the cart a user is linked directly to the source collection, from which the specimen(s) can be requested. Many databases of individual microbial collections are interoperable due to the efforts of projects such as the Common Access to Biological Resources and
___________________
6 See https://dwc.tdwg.org.
7 See https://www.tdwg.org/standards/abcd.
8 See http://gcm.wfcc.info.
Information consortium9 or the now-defunct StrainInfo (Verslyppe et al., 2014), which helped build common datasets based on specific data standards and formats.
Cyberinfrastructure in Support of Biological Collections
Biological collections may offer solutions to various major societal challenges relating to biology and the environment, from the emergence of new pathogens or the need for new antibiotics to the response of species to climate change, but this is possible only if the data can be accessed, aggregated, and analyzed effectively (Cook et al., 2020; Fontaine et al., 2012; Rocha et al., 2014). Following FAIR data principles (i.e., data that are findable, accessible, interoperable, and reusable [Wilkinson et al., 2016])—and the TRUST principles for digital repositories (i.e., repositories that promote the principles of transparency, responsibility, user focus, sustainability, and technology [Lin et al., 2020]) will require a robust cyberinfrastructure. As the digitization of biological collections continues to create large and diverse datasets, an effective cyberinfrastructure will need to incorporate mechanisms to improve access to an ecosystem of digital repositories and enable the integration of diverse types of data. Recognizing the need for a more robust cyberinfrastructure, the Earth science community established EarthCube in 2011 with NSF funding from both the Directorate for Geosciences and the Office of Advanced Cyberinfrastructure of the Computer and Information Science and Engineering Directorate at NSF.10 Collaborative projects with the biological collections community (such as enhancing Paleontological and Neontological Data Discovery API11 and Earth-Life Consortium12) as well as products resulting from EarthCube have been recommended for adoption by the biological collections community (e.g., Hobern et al., 2019). A similarly broad, community-level endeavor has not yet taken place between the biological science and computer science communities, but the timing is right, given the past decade of focused digitization.
For any local digitization effort to be successful, individual collection-holding institutions need a basic desktop computer and access to server infrastructure in order to house collection management system (CMS) databases, image repositories, and the necessary software for data publishing. Collections also require a workforce skilled in data management as well as collections curation and taxonomy. Both the natural history and living collections communities are using a large number of unique CMS databases that range from simple spreadsheets to more sophisticated systems that allow for database management and data manipulation, such as feature-rich SQL or Oracle-based systems (Arctos, Collections Space, Specify, BRAHMS, Axiell EMu, BioloMICS, GRIN, etc.) with extensive data models, collection management, and publishing capabilities.
___________________
9 See http://www.cabri.org.
10 See https://www.earthcube.org/info/about.
11 See https://www.earthcube.org/group/epandda.
12 See https://www.earthcube.org/group/earth-life-consortium-elc.
Data publishing increases the discovery of specimens for traditional research uses, for research that makes use of the digital data themselves (e.g., predictive modeling, recording of traits through optical character recognition of textual notes, or by machine learning from images), for formal and informal education, and for other novel downstream uses. While many institutions do not have the resources in house to install and maintain the necessary cyberinfrastructure to run a collections database and make their data available online, hosting services provided by web-based collection management packages and community-based solutions provide the cyberinfrastructure and technical expertise necessary to facilitate the digitization and publishing of these collections.
CHALLENGES
Realizing the promise of the digitization revolution will require overcoming a number of challenges. On one hand, there is an extensive community-wide backlog of specimens and associated materials that need to be digitized, creating gaps in our knowledge about the world’s biodiversity and missed collaboration opportunities between researchers. On the other hand, the multiplication of shared databases that vary in data quality and format and the proliferation of data aggregators and repositories can lead to an unnecessary duplication of effort, data disintegration, and limited data usability. Mass digitization is exposing digital data to an ever-increasing diversity of users for a myriad of uses, resulting in an increasingly complex digital landscape. Addressing these challenges will require the development, support, and maintenance of robust and coordinated cyberinfrastructure that provides for the ever-increasing needs of the world’s biological collections.
Dark Data
While the majority of data generated today are immediately digitally captured, historical collections typically have a backlog of data that have yet to be digitized. The digital revolution and the increase in the accessibility of digitized specimen data have been so profound that undigitized collections are now referred to as “dark data”—referring to the fact that they are essentially unavailable for modern scientific study without physical access to the specimens within institutions (Heidorn, 2008). The absence of these specimens from the global and national collections digital infrastructure represents lost opportunities for research and education as well as limits to returns on the investments made by the funding agencies that supported the acquisition of the specimens, even if the research projects that generated the undigitized collections were otherwise successful.
Discipline-Specific Limitations and Biases
Although digitization efforts to date have been transformational for both biological collections and research communities, most U.S. specimens, especially
those from taxonomically diverse groups, remain undigitized and unavailable for inclusion in cutting-edge research. The process of digitization can be particularly challenging for some disciplines where specimen labels are obscured or scarce, where taxonomic diversity is high and poorly known, where the type of preservation precludes automated capture of information (wet specimens in alcohol, for instance), or where the availability of historical paper records (card catalogs, ledgers, field notes, etc.) is limited. For example, for natural history collections, it is estimated that more than 50 percent of vertebrate collections (Krishtalka et al., 2016) and 20 percent of herbarium specimens (Barbara Thiers, Director of the William and Lynda Steere Herbarium at the New York Botanical Garden, personal communication, 2020) are digitized and available online, while only 4 percent of entomology collections have been digitized (Cobb et al., 2019), and most invertebrate biodiversity remains unknown or ignored (Di Marco et al., 2017). Plaguing biodiversity research, taxonomic bias13 also leads to a disproportional amount of dark data for certain collections and resulting discrepancies in knowledge from organism to organism across a wide range of biological fields (Adam et al., 2017; Clark et al., 2002). Multiple logistical and technical factors contribute to this bias, such as those mentioned above, but regulatory bottlenecks and restrictions play a role as well. Large-scale digitization efforts reveal the extent of century-long sampling and taxonomic limitations and biases and provide insights on how to account for such issues to inform future collecting (Daru et al., 2018; Troudet et al., 2017) and digitization efforts. For some biological collections, certain data fields need to be redacted or restricted and kept dark to protect sensitive information or specimens. This might include the exact geographic location of an endangered orchid or a fossil site on federal land, information and access to particularly virulent strains of biothreat pathogens, and personal identifiers in the case of organisms or samples originating from human specimens.
Project-Based Collections
A potentially large body of dark data lies in project-based collections—a group of specimens or samples collected with a particular purpose (e.g., for a specific research program, project, or survey of a group of organisms in a particular region) but never transferred to a permanent physical repository (e.g., museum collection or biological research center). While these valuable collections could make important contributions to science and society, the key problem is that they typically reside in an investigator’s lab, freezer, or office, making them difficult to identify and locate (for more, see Chapter 4). Typically, these collections are not accessioned, digitized, and made accessible to the wider scientific community through national data portals or catalogs. The barriers preventing accessioning into repositories and the subsequent digitization can be diverse. While some projects produce scientific publications that describe their findings and the materials accumulated, researchers may not be willing to share—or may be
___________________
13 The fact that some taxa are more investigated than others.
reluctant to relinquish control of—the specimens in their project-based research and thus be hesitant to contribute them to a publicly available repository or data portal. Even when researchers are willing to contribute their specimens and data, sometimes collections simply do not have the capacity or the resources to entertain such requests because of limited space and inadequate funds for accessioning and digitizing the specimens. Some project-based collections may not be suitable for incorporation into a permanent collection or digitization because of the recipient institution’s acquisition policies and guidelines (e.g., strategic growth, accessioning limitations, permits) or an inability to assess the value of a project-based collection and its benefit to the institution.
Private collections are also difficult to find. While outside the purview of this report, these private collections may hold essential data for documenting biodiversity, which may eventually be accessioned in public collections. Although the number and holdings of private collections in the United States are unknown, a recent survey in Europe found that private collections there may make up as many as 33 million specimens (Willemse et al., 2019). There are obvious issues concerning data quality and the willingness of these private collection holders to digitize and publish the data associated with the collections, but this information from Europe suggests that U.S. private collections may be a particularly valuable source of biodiversity data currently invisible to the research and education communities.
An Inefficient Data Pipeline
Currently, each online portal or aggregator collects a copy of a collection’s data published on a local database and ingests, normalizes, aggregates, and re-publishes this copy online. However, the current data publishing landscape lacks a streamlined and standardized pathway for carrying out these steps. For instance, if a collection shares its data with multiple aggregators, each aggregator may serve slightly different versions of the same record because they each have different publication schedules and different displayed fields for the specimen data. This publishing process and subsequent data verification steps (taxonomic and geographic verification, data cleanup, annotation, etc.) result in a massive duplication of effort by the aggregators and they each reconcile the specimen digital data while also creating confusion on the part of data users presented with multiple, yet slightly different, copies of the same data. Thus, while large amounts of data are appearing in portals, effective access to these data requires informatics expertise to remove duplicates prior to research use. As a consequence, some researchers and educators who may lack sufficient data management skills will rely solely on a single portal rather than exploring other portals for additional data—a practice that likely limits the number and possibly the diversity of the specimens obtained from a search. Furthermore, there is no effective mechanism in the current data publishing model for effectively and efficiently returning user annotations of data to the original data providers for incorporation into the data stream, resulting in a complete loss of this effort on the part of users of the data for the collections community. Leading aggregators such as GBIF, iDigBio, GCM,
ALA, and others recognize the problems of duplicate records and version control (Hobern et al., 2019) as well as the inadequate methods for annotation, but so far they have been unable to develop either a short-term fix or long-term solutions.
Variability in Data Quality and Format
As the quantity of digital data dramatically increases, the presence of incomplete data, data of questionable quality, and a lack of standardization limit both the roles that biological collections data can play in research and education and their usefulness. Issues such as incomplete data records and inaccurate or poorly transcribed data are ubiquitous and lead to limitations on the use of specimen digital data. For instance, an investigator searching on higher-level taxonomy, such as plant family, would miss records for which this information has not been recorded at a higher level but only at a lower one. Studies attempting to quantify the timing of animal migration or plant flowering would be severely hampered by a lack of specific temporal information. Some disciplines (e.g., botany) have used skeletal records14 as an initial step in digitizing specimen records in order to save time (Nelson et al., 2015; Rabeler, 2015), but while this method opens up a large number of records for discovery, some of the information in these records has yet to be completely digitized, meaning that certain fields of information are not readily available for research. Similarly, although some disciplines have made great strides in community georeferencing15 endeavors, such as the NSF-funded Mammal Networked Information System, ORNIS (A Community Effort to Build an Integrated, Distributed, Enriched, and Error-checked ORNithological Information System), HerpNet, and Fishnet collaborative projects (Chapman and Wieczorek, 2006), many specimen records are not yet georeferenced and are thus unavailable for spatial analyses such as ecological niche modeling and species distribution analyses (Bloom et al., 2017; Seltmann et al., 2018). Other locality records may never be able to be georeferenced because of historical limitations in the precisions of their locality information.
Data transcription errors and a suite of taxonomic naming issues (Nekola et al., 2019) create a variety of other issues. For example, the rate of errors in geospatial designation or taxonomic classification, either through synonymy or misidentification, has been estimated to range anywhere from 5 percent to 60 percent (e.g., Goodwin et al., 2015; Nekola et al., 2019). Without adequate taxonomic resolution, taxonomic incongruencies can result in incomplete species distribution and trait information. In addition, a lack of adherence to standardized terminology and controlled vocabularies, as well as limitations of or incorrect mappings to Darwin Core fields, has led to various problems in data analysis. For example, attempts to compile information on all “females” of a species are hampered by the numerous variants of this term in the sex field—F, Female, female, etc. (e.g., 2,800 distinct values appear in the sex field in VertNet; see Guralnick et al., 2016). Approaching the issue at the source by standardizing
___________________
14 A basic set of data per specimen (Nelson et al., 2015).
15 The process of converting a text-based description into a geospatial coordinate.
and controlling vocabulary in local collections databases and providing common names for organisms would increase usability, but a consensus on taxonomy, terminology, and common names among scientists, which will be needed in order to enable such functions, is still elusive in some disciplines.
Limitations Affecting Data Usability
Once published to a portal, digital datasets require collections professionals to curate and maintain their quality, just as physical specimens require specialized care. Inadequate maintenance of these datasets can severely impair the use, value, and impact of biological collections data in research and education. Both local and community-level mechanisms could improve the quality of their data. One challenge is the lack of expertise by collections professionals in evaluating data quality across broad taxonomic distances and types of data, although standardized vocabularies could provide the necessary tools to assess data completeness, quality, and consistency and to increase the fitness-for-use of biodiversity data (Ball-Damerow et al., 2019). Data transcription errors also require correction by individual collections or potentially through community efforts (see Nekola et al., 2019, for a summary). However, digital datasets are often not maintained and updated for a variety of reasons, ranging from insufficient resources and staff turnover to disputes related to intellectual property rights and to a simple lack of understanding that digital datasets are not static, one-off products.
Another factor affecting data usability is the fact that data portals have been developed for different uses and different communities and their interfaces are not always user-friendly for either the public or the research community. Their design has often been an afterthought because the interfaces for most portals are designed with a single purpose in mind and anticipate only one type of user—the research or collections specialist, and not the general public or student users (Hendy and MacFadden, 2014). Thus, although millions of specimen records are available online, the level of technical expertise necessary for accessing them may be too high for some users. Portals that were designed to serve a wide array of data (e.g., GCM and GBIF) also suffer from limited search capability. Fields that are unique to particular collection types (e.g., mutant allele for genetic stock centers or geological data for paleontological specimens) are not searchable, making those data more difficult to discover. Currently, many data portals are available only in a single or a few languages, providing yet another barrier to accessibility and contribution.
Inadequate Methods for Data Integration and Attribution
Realizing the vision of successfully integrating and tracking data from various sources carries many challenges, most significant of which are issues of scale and interoperability. Data integration relies on the unambiguous identification of individual data elements, packets of data, and people through the use of globally unique identifiers (GUIDs), digital object identifiers (DOIs), and open researcher and contributor IDs (ORCIDs) (Page, 2008) as well as the implementation of standardized application programming interfaces and
exchange formats (Konig et al., 2019). Despite several attempts (e.g., Güntsch et al., 2017; Guralnick et al., 2014; Nelson et al., 2018), the biological collections community has been unable to agree on a single identifier to describe data elements, though many candidates have been proposed (GUIDs, life science identifiers, uniform resource identifiers, DOIs, Darwin core triplets16). Although most collections now use some form of identifier as listed above, there is no centralized system of registration to ensure the uniqueness—and therefore traceability—of these identifiers, and attempts to link data informatically have been only marginally successful (e.g., Guralnick et al., 2014). Because living stock and natural history collections databases were established in parallel using different types of identifiers, integrating them has proved to be quite complex, and these difficulties may preclude opportunities to integrate the data from these resources. The challenge is exacerbated by the differing types of published data not being comparable, by differing expertise, and by the different user communities being served. As a result, tracking the use of biological collections data in research and education still remains largely a manual and time-consuming endeavor. Issues of tracking multiple identifiers and integrating specimen data across databases and portals are exacerbated by the fact that identifiers do not reliably persist through to the products of research created from the use of these specimens (Arbeláez-Cortés et al., 2017; Rouhan et al., 2017). In fact, even the way that specimens are cited in published work is inconsistent, if they are cited at all. This results in a lack of recognition and attribution of the contribution of biological collections to research and education.
Despite all of the challenges described above, electronic citation and tracking of digital specimen records, each with a unique identifier, can provide attribution to local collections and can enable assessment of short- and long-term impact, both locally and nationally. Although digitization of biological collections has provided access to massive numbers of specimen records, the assessment of the impact of this resource has barely begun (Hobern et al., 2019; Lendemer et al., 2020). Few biological collections have the resources or community-based guidance to take the next step in determining the contributions of their collections to the published scientific body of knowledge. For example, due to incomplete or non-unique metadata in GenBank, even the apparently simple task of automatically connecting genetic data from GenBank to voucher specimens in iDigBio cannot be accurately accomplished, although this connection may be established manually for a given collection, as demonstrated more than a decade ago (Strasser, 2008). While technology may offer some solutions, the development of such citation and attribution systems is in the early stages of implementation—see occCite17 and GBIF citation metrics and guidelines18 as promising examples—and it will require substantial investment if these are to be implemented on large scales. The problem is compounded by a lack of coordination among the members of the biological collections community and
___________________
16 A concatenation of values for institution code, collection code, and catalog number for a specimen.
17 occCite is an online tool that enables biological collections to track how their data are being used. See https://hannahlowens.github.io/occCite.
by a lack of appropriate resources to develop and implement an assessment of collective impact. Investing in the development of bioinformatics tools and cyberinfrastructure to capture data used in publications and other forms of output could be transformational in making it possible to accumulate national usage statistics and to carry out rigorous evaluations of the impact of both physical and digital resources.
Limited Mechanisms to Support a Cyberinfrastructure That Promotes Collaboration
The diversity of biological collections poses many challenges to the effective development and implementation of a cohesive, adaptable, and sustainable cyberinfrastructure that serves the entire collections community. For example, inherent differences between living and natural history collections such as differing needs and goals, compounded by external factors such as different funding opportunities and requirements, have thwarted collaborative efforts to integrate digital data from these collections. Many natural history institutions with the necessary funding for personnel and technology have been digitizing their collections for four decades (Nelson et al., 2018), but NSF’s 10-year, $100 million ADBC program, launched in 2011, has led to even greater strides in digitization and provided access to an ever-increasing quantity of data from natural history collections. In contrast, at present, living stock collections are ineligible for funding through the ADBC program, and, for now, no comparable programs specifically fund the digitization of living biodiversity collections. The immense amount of digital information being produced by current digitization efforts and the data integration challenges outlined above threaten to outstrip the necessary cyberinfrastructure support (storage devices, backup systems, routine maintenance, and technological upgrades). The financial outlay required for these necessary components and additional workforce needs (see Chapter 6) is sometimes not adequately factored into the cost estimates of digitization, so that the infrastructure components and workforce needs are left unfunded (see Chapter 7), with it being necessary to put retroactive measures in place to address the issue in hindsight. Without sufficient investment in these cyberinfrastructure components and support by individual collections, funders, and the community as a whole, the amount of digital data stored, shared, and integrated will continue to be limited for certain collections. However, it is precisely a broadly based, flexible, and robust cyberinfrastructure that could integrate complementary data from living and natural history collections (e.g., microbiome studies, food safety, biotechnology applications, etc.) or other groups of collections.
THE WAY FORWARD
Digitization is increasing the relevance of collections in diverse ways and allowing collections around the world to network their way toward the “global museum” that will seamlessly integrate worldwide collections (Bakker et al.,
2020). To date, the digitization of biological collections has proved extremely valuable and successful. The result has been new partnerships for innovative scientific inquiry and learning. Digitization has significantly increased the accessibility and usability of biological collections data for traditional research, for new research of global societal importance, for education (e.g., Cook et al., 2014; Powers et al., 2014), and for an ever increasing and ever more diverse collection of additional applications (for review, see Ball-Damerow et al., 2019; James et al., 2018; Krishtalka et al., 2016; Nelson and Ellis, 2018).
However, if such successes are to continue and multiply, a great deal of work remains to be done. A large percentage of the nation’s biological collections have not yet been digitized. Data cleaning exercises, standardization, and the provision of annotation mechanisms will significantly increase the usefulness of both the collections that have already been digitized and those that will be digitized in coming years. Finally, digitized biological collections will be most valuable as components of a highly integrated cyberinfrastructure that provides easy access to the collections, integration among different collections and with data beyond collections (such as environmental data, genetic data, and biodiversity analyses), and a way to enable effective collaboration among the many researchers who work with those collections and among potential users of the data. These steps will make it feasible to fulfill the extraordinary promise of digitized biological collections.
Innovative Approaches to Reducing Dark Data
Given the foundational role that digitization plays in the development of an accessible, useable, and networked scientific infrastructure, it is important that biological collections continue to digitize and to provide data that are of high quality, in a standardized format, fit for use, and broadly accessible. Digitization workflows are currently in place in many communities and institutions, and systematic digitization is set to become more efficient than in the past thanks to ongoing training support by iDigBio and others. The quantity of digital data available for end use is determined not just by the pace at which historical data can be digitized, but also by the efficiency of adding new field-collected materials or project-based collections to permanent repositories and online portals. To avoid contributing to the backlog of undigitized material, the large amount of data associated with these new specimens needs to be “born digital.” Streamlining their integration into collection databases and online data aggregators will require collaboration among field collectors, collections professionals, and the informatics community. By building on recent achievements of the collections community, future efforts to digitize most U.S. collections seem feasible, given sufficient time and funding.
Massive digitization efforts to capture and place online not only the metadata associated with biological specimens but also high-resolution images of the specimens themselves, along with videos and vocalizations, have unleashed entirely new areas of study. Thanks to new imaging techniques and technologies, the use of rare or fragile natural history collections is less invasive, and it is possible to carry out detailed examinations of specimen attributes without extensive
handling of the specimens themselves (see Box 5-1). Sensitive computed tomography (CT) methods of scanning whole organisms and individual skeletal elements capture anatomical features in unprecedented detail and permit precise three-dimensional replication of specimen morphology. Other technological advances have made the digitization of some collection types less time consuming and more efficient (e.g., trays of insects with multiple labels, fluid-preserved specimens, microscopic organisms). Batch processing or automation and the use of optical character recognition (OCR) have shown some success in optimizing the capture of text from specimen label images. The secondary augmentation
of records through georeferencing19 can be facilitated through the use of online software such as GEOLocate.20 Specially designed robotic systems that select and image individual specimens or scan whole drawers of specimens and their data are now a reality. The use of convolutional neural networks, a form of machine learning that has been used for species identification (e.g., Carranza-Rojas et al., 2017) and the capture of trait information from specimen images and text such as whether a specimen is in flower or fruit (e.g., Lorieul et al., 2019), is another
___________________
19 Assigning a latitude and longitude to a collection locality (e.g., GEOLocate, Google Earth).
area of innovation that could advance digitization (see Box 5-2) and that is ripe for collaboration with computer scientists.
The natural history collections community has begun to use outside assistance in the digitization process in an effort to reduce the amount of dark data. The impact and contribution of citizen scientists and volunteers to the digitization effort have steadily increased through efforts such as Notes from Nature21 and the Smithsonian Transcription Center,22 among others. The annual Worldwide Engagement for Digitizing Biocollections (WeDigBio)23 global transcription event has also galvanized these digitization efforts by engaging a large and diverse set of individuals from varying backgrounds in the digitization process (Ellwood et al., 2018). Although these citizen science efforts were originally designed to assist with the transcription of specimen label data, field notes, and other text (Hill et al., 2012), citizen scientists are extending their contributions to other forms of data capture, such as scoring herbarium specimens for phenological phase. Despite lingering skepticism about the quality of data produced by citizen scientists, it has been found that, when given appropriate instructions, citizen scientists produce data that are on par with specialists (Brenskelle et al., 2020; Catlin-Groves, 2012), and the power of engaging citizen scientists is shown by the fact that the 4-day WeDigBio event in 2018 resulted in more than 50,000 record transcriptions (Ellwood et al., 2018). However, despite the addition of these efforts to existing collections digitization efforts, most of the nation’s collections remain to be digitized.
It is important to note that the physical specimen is the nexus for the digitized data associated with it and that it should not be neglected or discarded. Often, the specimens remain the primary source of verifiable biodiversity data, and the curation of the underlying specimens required for such analyses remains paramount, especially if researchers want to later examine the physical specimens after analyzing data from digitized information such as images or genetic sequences. For example, downstream analyses can include the extraction of DNA for the confirmation of species identifications based on analyses of digitized specimens or a simple inspection of specimens for verification and occurrence that might appear anomalous in terms of locality or habitat. As such, digitization is not a substitute for physical specimens, but rather a necessary complementary activity that exponentially increases the usefulness of and provides wider access to the collections of these physical specimens. In fact, evidence is accumulating that use of physical specimens through loans and visits to collections has actually increased with the recent online accessibility of digital records (Vollmar et al., 2010). For living stock collections, continued digitization allows researchers around the globe to locate and acquire an ever-growing number of existing and newly developed model organisms, with the digital data being more of a finding tool and the physical specimen still remaining vitally important. In some cases, such as in the case of destructive sampling or loss of a specimen, the electronic
___________________
21 See https://www.notesfromnature.org.
22 See https://transcription.si.edu.
23 See https://wedigbio.org.
information stored in a database becomes the only record available; this is the case especially for a growing number of microorganism specimens (see Box 5-3), and thus digitization is essential for future studies that aim to understand their biology and evolution.
Increasing Data Visibility
Although digitization and sharing data with online open access data portals continue to provide more data for research and education, vast amounts of data produced through research and collecting endeavors, such as project-based collections data, are still not publicly available. This is particularly prevalent at institutions that lack permanent collections. Making these data public would increase the visibility of the data as well as promote research reproducibility and reduce redundancy. The primary onus of ensuring that data are captured
and disseminated falls on funding agencies, reviewers, and publishers. The NSF Directorate for Biological Sciences requires a data management plan as part of all research proposals, but while this is a prerequisite for funding for living stock collections, there is no requirement for digitization, publishing, or ensuring the long-term accessibility of specimens and their data for natural history collections. There is thus an opportunity to develop more stringent requirements for managing and archiving specimens and their data as part of a specimen management plan (see also Chapters 4 and 7). Likewise, there is no uniformity in the requirements for data citation in publications through journals. Publishing entities along with their editorial boards (and with pressure from funding agencies) could enact
uniform requirements for data citations in order to promote reproducible science as well as to provide the necessary mechanisms for collection attribution.
Tools to Improve Data Quality
Emerging efforts to provide online tools for improving data quality while also facilitating data integration, usability, and accessibility to a broader range of communities hold significant promise in many areas. Both discipline-specific efforts to address data quality and larger-scale efforts by data aggregators provide such opportunities. The aggregator community has a major role to play, with GBIF, iDigBio, ALA, GenBank, and VertNet having already incorporated data quality tests and assertions24 into their portals, which, in some cases, automatically correct or augment records to enhance their fitness for use (Bouadjenek et al., 2019; Chapman et al., 2020). Most of the changes made as a result of these tests and assertions improve data quality by identifying georeferencing mismatches, genetic sequences that are inconsistent with the literature, taxonomic or geographic anomalies, duplicates, or issues related to data standards or vocabulary. Currently, there is no uniformity in the identification of the errors or in the implementation of the edits across the various aggregators, but recommendations to improve data quality have been proposed (Chapman et al., 2020; Groom et al., 2019). In addition, there is a need to create standardized and consistent mechanisms for feeding these corrections and data flags, or annotations created by users of the data, back to the data providers in order to inform data correction and augmentation at the source. In some cases, annotations and errors found by the users of the data are provided to the data providers in a format requiring corrections to be made individually, one record at a time, which is simply not feasible for large datasets. In the past few years many web annotation tools for eliminating these hurdles have become available (Suhrbier et al., 2017; Tschöpe et al., 2013). Partnering with computer scientists and software developers could lead to the deployment of mechanisms for routing data quality annotations to the data providers and for those annotations to be easily reviewed and integrated into the source data in batches. Machine learning and other forms of artificial intelligence may provide incremental increases in the annotation of certain collections, primarily through text recognition and OCR technologies using images of labels or card catalogs or ledgers. A systematic and standardized approach to improve data quality will result in optimized user experience. Some portals have started to adopt the use of facets, filters, or auto-complete for searching, rather than completely blank entry fields. Such modifications are also steps in increasing the accessibility of collections data to a wider range of users.
Promoting Integration and Attribution
Many national and international organizations have developed standards for collections data management that inform the integrity and format of digitized
___________________
24 A query that looks for problems in a biological collection dataset.
data. These data associated with specimens usually involve a suite of unique identifiers with taxonomic, locality, temporal, and preparation information as well as various collections management–based fields (catalog number, cataloger, etc.). While the fields of information captured may vary by discipline or collection, the widespread adoption of GUIDs would allow for a much deeper and broader integration of data both within and among collections. Collections with a critical body of digitized data based on or derived from the specimens are now interested in linking their basic collection metadata to information such as gene sequences, isotope values, or morphometric analyses. Such linkage will further improve data integration and create better connections between primary specimen records and extended data. Linking the data in this way creates what has become known as the “extended specimen” (Webster, 2017) (see Figure 1-2). Extending specimen data with these resources greatly increases the value of the digitized collection for downstream uses while promoting integrated science (Lendemer et al., 2020; Thiers et al., 2019). A lack of integrated online resources will restrict access to valuable collections information, limiting the uses of the data in research and the potential scientific discoveries related to those data. For maximum use, digital data require integration and interoperability at multiple levels. At the specimen level, data derived from the diverse preparations of each specimen (e.g., skeletons, tissues, parasites, field notes, publications, etc.) need to be connected in order to create full extended or holistic specimens for multidisciplinary applications. In addition, these data need to be integrated with the new data streams derived from subsequent investigations (e.g., GenBank sequences, IsoBank signatures, images, CT scans, viromes, and various -omic data). At the collection level, creating associations among taxonomically disparate specimens to highlight such relationships as tissue–voucher, host–parasite, pollinator–host plant, predator–prey, commensals, and others are crucial for integrative science. At the ecosystem level, many novel uses of biological collections data, such as evaluating species’ responses to global change, require integration with other forms of data, such as genetic, observational, trait, environmental, geographic, ecological, and remote sensing data. Such an integration will not only require the collections to be more robust and complete but will also necessitate the creation of interoperable linkages among databases. Some levels of integration of disparate datasets are currently being achieved on a national and global scale through various aggregators and individual museum data management systems, but more coordination between these aggregators and developers is needed to simplify and standardize the landscape.
A cyberinfrastructure for biological collections could enable data integration while also providing annotation tools and a system for attribution of specimen data used in research, education, policy development, or other activities of this scope. Creating such a cyberinfrastructure will require robust technological cyberinfrastructure tools to link data elements and also social incentives that will engage all actors in the data pipeline from collections, to researchers, aggregators, data authority providers (taxonomy), journal editors, and beyond. The promises of data integration and attribution were addressed in a Biodiversity
Collections Network workshop (BCoN, 2018) in which a possible system of identifiers and linkage mechanisms was identified as a solution to better integration and attribution of digitized biodiversity data. For example, a number of systems that are intended to solve various aspects of the integration process are being developed (e.g., GenBank Linkout25 and Pensoft ARPHA writing tool26), but while there are analogous systems in other domains that one could learn from or co-opt (e.g., research resource identifiers of the Resource Identification Initiative27), no comprehensive solution has been forthcoming. The more that such technological solutions are implemented, the less the community will need to rely on social solutions where all producers and users of data need to perform linkages manually. The broadened utility of collections data, through integration with other data sources, will eventually increase the use of collections (both physical and digital) and thereby increase the attribution, tracking, assessment of impact, and subsequent advocacy for these resources. Assigning identifiers to downloaded datasets from aggregators would also promote both attribution of data use to the providing institution and reproducible science. For example, GBIF assigns a DOI for a downloaded dataset, but recent research has shown that neither URLs nor DOIs are stable, even over short timeframes, and suggests instead a method of cryptographic content-based identifiers (Elliott et al., 2020). Continued efforts to develop methods for identifiers of datasets to enable data integration, attribution, and reproducible science are needed. One technological solution that could potentially resolve the data integration and attribution problem and that has recently received attention is blockchain (van Rossum, 2017). Blockchain is used most commonly in cryptocurrency where it provides an incorruptible digital ledger of economic transactions. A blockchain-inspired network has the necessary technological components to provide the identification of the various elements of the network while also tracking all transactions associated with each item. The network could take advantage of the existing identifiers commonly used in the collections community (GUIDs, DOIs, ORCIDs, etc.) to effectively identify occurrence records, data downloads, publications, and agents. Transactions such as a change or augmentation of the record by the collection, an aggregator or a user of the collection, a loan or gift of material by a collection to an end user, the lodging of a DNA sequence with GenBank, or the publication of results depending on the use of physical specimens or data could all be digitally recorded by the blockchain network. Each of these individual transactions would be logged by the system and would be traceable and immutable. Further investigation of a blockchain-based cyberinfrastructure could yield innovations for managing and tracking all activities of biological collections.
___________________
25 See https://www.ncbi.nlm.nih.gov/projects/linkout.
26 See https://arpha.pensoft.net.
27 See https://www.force11.org/group/resource-identification-initiative.
Developing a National Cyberinfrastructure
As digitization spreads across scientific disciplines and data sharing becomes more common, the development of a flexible, unified, and sustained national cyberinfrastructure would provide greater opportunities to integrate and support disparate digital datasets such as living stock and natural history collections and would facilitate research and educational opportunities. This shared resource would not only serve the needs of the collections communities but also provide a baseline to all biodiversity knowledge. Partnerships and pooled resources may be the key to the development and implementation of a permanent, effective cyberinfrastructure in support of digitization, annotation, integration, and analysis of the nation’s collections. Because small collections may have unique holdings that reflect regional species pools or the expertise of present and past local collectors and researchers, making these collections digitally available will be a first step toward greater advocacy, visibility, use, and inclusion in large-scale studies. However, some small collections do not have the resources to manage their own cyberinfrastructure or establish and maintain a portal to store their data or even publish them online. The cyberinfrastructure needs of these collections are in some cases being addressed at the community level through cloud hosting of collections databases (e.g., Arctos,28 Specify,29 or BioAware30). These web-based collection management packages offer information technology support, which is often not provided in-house by the institution but is necessary to facilitate digitization and publishing of collections. This model has the additional benefits of making data publishing streamlined and making connections to external data repositories more robust (e.g., GenBank, Morphbank, IsoBank, Morphosource, Ontobrowser, DataOne). Data portals such as iDigBio provide global access to digital data from U.S. collections and are therefore a key feature of cyberinfrastructure, but they in turn rely on additional cyberinfrastructure components, such as hardware for servers and storage, an evolving database schema to accommodate innovations in digitization, and a workforce capable of adapting to a rapidly changing data science landscape (see Chapter 6). This type of infrastructure needs to be maintained at the national level, for use by and the benefit of the biological collections community as a whole (see Chapter 8).
A Robust Cyberinfrastructure to Promote Coordination and Collaboration
Connecting data in order to generate shared resources has additional benefits. For example, researchers are increasingly interested in patterns of spatial, environmental, and genetic variation, particularly when evaluating how species might respond to climate change. Data from living stock and natural history collections, environmental databases, the National Ecological Observatory Network, and GenBank would all contribute to addressing these questions, and
___________________
28 See https://arctosdb.org/about.
a cyberinfrastructure to support these linkages would enable important new science while ultimately reducing costs through the elimination of duplicated effort. Moreover, the development and deployment of analytical tools and pipelines through unified resources would democratize biodiversity science by allowing accomplished biological specialists who are not well trained in informatics and computer science to address important basic and applied research. Collaboration with a national cyberinfrastructure for life science research, such as CyVerse31 (funded by the NSF Directorate for Biological Sciences), the Texas Advanced Computing Center,32 the Extreme Science and Engineering Discovery Environment,33 and the Data Observation Network for Earth,34 could provide resources to support biological collections and lead to an enhanced national network of digital data from collections and other relevant repositories by improving accessibility to and linkages among data from different sources. The EarthCube community (see above) could serve as a model for how such a collaboration might be implemented.
A national cyberinfrastructure for biological collections that will support these collections and facilitate their ever-growing base of end users will require collaboration, especially between the collections community and computer scientists and engineers, but also between collections staff from diverse collections types and communities (e.g., natural history and living stocks). Until recently, interactions between these communities have been limited due to a lack of funding and staff availability (see Chapters 6 and 7). However, the effective development and deployment of cyberinfrastructure for biological collections will require both (1) application of recent advances from computer science and engineering in new contexts and (2) innovation of cyberinfrastructure components to meet the unique needs of biological collections and an ever-widening user community (e.g., Heberling et al., 2019). Successful implementation will require an interdisciplinarity that is only beginning to emerge among computer and data science and all fields of biology represented by biological collections. To date, innovations in the development of the world’s largest aggregators of data from natural history collections (e.g., GBIF, iDigBio, ALA) and living collections (e.g., GCM) have resulted from close collaborations among biologists, data scientists, and engineers. Moreover, because some computer and data scientists are embracing the data from these biological collections (Chen et al., 2019; Drew et al., 2017), interesting challenges for machine learning and analytical pipelines are being tackled. A similar, although perhaps less appreciated, facet of the situation is that biological collections provide unique and scientifically interesting challenges that could possibly benefit the computer science community, perhaps with extensions to problems outside of collections. NSF’s Big Idea “Harnessing the Data Revolution” is certainly relevant to collections data, particularly as both the volume and heterogeneity of data increase and as researchers and
___________________
31 See https://www.cyverse.org.
32 See https://www.tacc.utexas.edu/-/tacc-a-holistic-approach-to-making-cyberinfrastructureaccessible.
33 See https://www.xsede.org.
34 See https://www.dataone.org/working_groups/cyberinfrastructure.
educators are increasingly interested in connecting collections data with other data resources, from environmental to genomic data. However, continued progress and new advances will require expanded collaborations. Formal efforts to bring these groups together through, for example, workshops, shared funding, and other opportunities would reap large rewards for the design and extension of cyberinfrastructure in capturing the many elements of the extended specimen and aligning data resources in living and natural history collections.
CONCLUSIONS
Certain impediments will have to be overcome before the potential of a national cyberinfrastructure and the digitization it supports can be realized. Through varied programs past and present, NSF contributions to biological collections digitization and cyberinfrastructure have been critical in the United States. To be successful and sustainable, the digitization and development of a robust cyberinfrastructure will require continued support from NSF. Although digitization efforts have involved hundreds of collections, phylogenetic, geographic, temporal, and taxonomic gaps in digitization are evident. Harnessing the opportunity for data-driven discoveries and transdisciplinary collaboration will depend on a continuing effort to digitize new and existing biological collections using developed communities of practice (e.g., best practices and standards). Investment in the development of new technologies and cost-effective, high-throughput workflows for digitizing collections that, to date, have lagged—such as entomological collections—will enhance both the number of specimens and the taxonomic scope of digitized collections. Future digitization initiatives will need to be prioritized to address this disparity in order to ensure better representation of data from these underrepresented groups. In addition, the identification, assessment, and accessioning of legacy project-based collections could bring a large number of valuable specimens and their digitized records into the public domain and prevent the future accumulation of inaccessible collections that diminish NSF’s investment in their assembly and future use in research and education. Compounding these issues is the lack of resources or associated workforce (see Chapter 6) and also staff who may not realize the value of the collections once digitized. If these dark data can be made available, both the physical collections and their digital representations can be used in future research, contributing to the growing fabric of networked collections.
National and global portals and catalogs have made important contributions to the biological collections community by providing a platform with which to exchange and share data and promote standardization and consistency. Continual updating, augmenting, and improving digital data records using annotation tools and data assertions, for example, will greatly improve overall data quality and, in turn, lead to more comprehensive data integration and greater accessibility of digital data. However, mechanisms for data annotation and attribution require an interoperability of data and systems, which may be impeded by global indecision about the application of globally unique identifiers for specimen
records. In addition, despite some progress, integrated systems that enable the citation of data used in research publications and attribution to data providers are difficult to develop and will require an all-encompassing approach with social incentives and innovative technological solutions. These are not insurmountable problems, but it will be important to address them in the development of a comprehensive national cyberinfrastructure for the large-scale, long-term digitization and use of digitized data.
The integration of specimen data with other biological components as well as with data sources outside of the biological realm will require the implementation of a network of cyberinfrastructure resources not yet realized. Possible future collaborations are potentially unlimited, but computer scientists and the collections community will require mechanisms to bring them together and instruction on how to communicate across disciplinary barriers. Rapid developments during the past few years argue for the value of these collaborations. Just as innovations in digitization have resulted from partnerships between these communities, further collaborations, particularly in the application of machine learning, will lead to even greater progress in digitization, georeferencing, and data analysis. A unified cyberinfrastructure that connects all types of biological collections, such as living and natural history collections, could accelerate research and provide innovative educational opportunities. Moreover, a permanent national cyberinfrastructure that supports the needs noted above in terms of expanded digitization of dark data, improvement in data quality, and an increased accessibility to digital data would certainly spur data use. Without this resource, collections—both physical and digital—will continue to be underused.
RECOMMENDATIONS FOR THE NEXT STEPS
Recommendation 5-1: The leadership (managers and directors) of biological collections should provide the necessary mechanisms for staff to keep pace with advances in digitization and data management through training in digitization techniques and publishing of standardized quality data that can be efficiently integrated into portals.
Recommendation 5-2: Professional societies should initiate and cultivate opportunities for research collaborations within the biological collections community. These collaborations should include working with the computer and data science communities to promote the development and implementation of tools to build the cyberinfrastructure (e.g., data storage, annotation, integration, and accessibility to expand the use of biological collections to a broader range of stakeholders).
Recommendation 5-3: The National Science Foundation (NSF) Directorate for Biological Sciences should continue to provide funding for the digitization of biological collections and for the cyberinfrastructure to support both living and natural history collections. Specifically, the NSF Directorate for Biological Sciences should:
- partner with other directorates within NSF (e.g., physics, chemistry, computer science, and education) and other federal agencies and departments (e.g., the Department of Health and Human Services, the Department of Agriculture, the Food and Drug Administration, the Department of the Interior, the National Oceanic and Atmospheric Administration, the National Aeronautics and Space Administration, the Department of Energy, etc.);
- establish ongoing mechanisms for the biological collections community to meet, develop best practices, and work toward goals such as establishing and implementing unique identifiers, clear workflows, and standardized data pipelines; and
- promote and fund the development of a necessary national cyberinfrastructure, with appropriate tools, and technology to affect the efficient multi-layer integration of data and collections attribution.




























