Genome Editing with Precision and Accuracy
KRISHANU SAHA
University of Wisconsin–Madison
Editing the genetic code of living organisms with wordprocessinglike capabilities has been a goal of life scientists and engineers for decades. For biomedical applications, changing as little as a single base in the 3 billion bases of the human genome might cure disorders such as muscular dystrophy and cystic fibrosis. New genome editing tools are now capable of “one in a billion” specificity, leading to the prospect of new classes of gene and cell therapies. To illustrate both the excitement and the risks, I begin with a cautionary tale that made global headlines, before describing opportunities and challenges in efforts to develop genome editors for biomedical applications.
A CAUTIONARY TALE
In November 2018 Chinese biophysics researcher He Jiankui surprised the world by announcing the birth of two genome-edited babies, the so-called “CRISPR1 twins” (Cyranoski 2019). It was a momentous step: the intentional alteration of the human germline to produce changes transferred to the next generation.
But the editing outcomes were not as Dr. He expected (Ryder 2018). The Chinese team had injected CRISPR-associated protein 9 (Cas9) genome editing RNA and proteins into human embryos to modify the CCR5 gene, which encodes a receptor on the surface of immune cells for human immunodeficiency virus (HIV). CRISPR-Cas9 is a nuclease that can cut DNA, and the location of the cut can be programmed by the sequence of a single guide RNA (sgRNA).
___________________
1 CRISPR (clustered regularly interspaced short palindromic repeats) is a family of DNA sequences in the genomes of prokaryotic organisms such as bacteria.
The sgRNAs used by Dr. He targeted two locations in the CCR5 gene to delete 32 amino acids of the translated CCR5 protein. The deletion of the CCR5 gene was meant to destroy the ability of HIV to infect T cells and thus make the treated embryos resistant to HIV. But genomic analysis of the CRISPR twins indicated that the intended mutation was not achieved; instead, different insertions and deletions of DNA bases (termed indels) were generated in the CCR5 gene. These insertions and deletions are anticipated to make the twins more susceptible to influenza, with unknown effects on their susceptibility to HIV.
While there are many lessons to be learned from this experiment (Barrangou 2019; Jasanoff et al. 2019), it is a cautionary tale about using genome editing tools with poor precision. Critiques of the approach indicate suboptimal use of both Cas9 nucleases (there are protein-engineered, higher-fidelity variants) and sgRNAs (other target sequences could have been used) as well as questionable timing of the intervention (the proteins and RNA could have been introduced at a different stage of embryonic development).
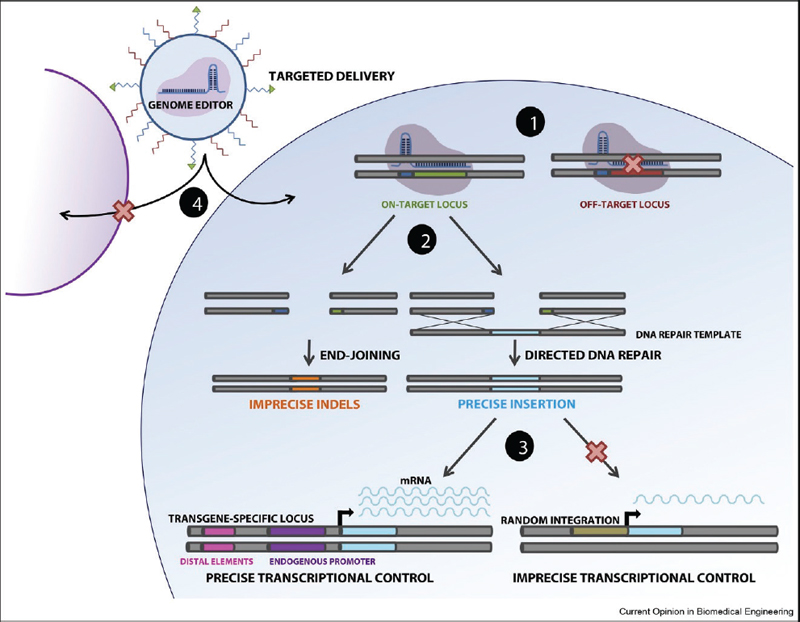
The challenges of getting all the parameters right are daunting, and many people, including leaders in the scientific community (Lander et al. 2019), argue that human embryo editing should not even proceed. There is intense activity, however, to attempt to tackle the challenges of editing the human genome after birth, so-called “somatic genome editing,” to mitigate certain diseases. I briefly describe efforts to overcome four types of challenges through improved precision and accuracy in genome editing (figure 1).
CHALLENGE 1: ON-TARGET NUCLEASE ACTIVITY
The most common interpretation of “precision” in genome editing is the ability to edit the genome at the intended target site while limiting edits elsewhere in the genome, commonly called “off-target effects.”
For the CRISPR-Cas9 genome editing systems, Cas9 has been observed to create excessive undesirable mutations (Cradick et al. 2013; Duan et al. 2014; Pattanayak et al. 2013). New methods have therefore been developed to controllably introduce genome editing components, such as ribonucleoproteins (RNPs), and to regulate when and where Cas9 is expressed (Chen et al. 2016; Davis et al. 2015; Hemphill et al. 2015). For example, modified Cas9 nucleases can be selectively activated with small molecules to decrease the gene editing time window (Davis et al. 2015).
Several groups have engineered nickase Cas9 proteins with only one active nuclease domain. A single nickase cannot create a full DNA double-strand break, but when two nickases are paired, the break can be repaired via nonhomologous end joining (NHEJ) in mammalian cells (Ran et al. 2013). While this method lowers off-target effects, the efficiency of the genome editing is greatly decreased as two nickases and two sgRNAs need to be delivered to the nucleus to perform simultaneous cuts.
Rational protein engineering approaches to modify the nuclease have also generated highfidelity variants of Cas9: eSpCas9 (Slaymaker et al. 2016), Cas9-HF1 (Kleinstiver et al. 2016), and xCas9 (Hu et al. 2018). Cas9 variants decrease the binding time of the sgRNA to the target sites in the genome, resulting in a decrease in off-target binding and cutting. These high-fidelity Cas9 variants may represent a quick path to clinical relevance as they can greatly reduce off-target events.
CHALLENGE 2: “SCARLESS” INCORPORATION OF NEW SEQUENCES
While the intended on-target gene can be modified using a targeted DNA break, attempting to insert specific bases (i.e., to “write in” sequences) into the genomic target site adds a layer of complexity.
DNA repair pathways in the cell dictate whether new nucleic acids can be inserted, and a major pathway used is the homology-directed repair (HDR) path-way. HDR can generate perfect incorporation of the desired sequence without modifying any other bases in the genome and so is called “scarless editing.” Researchers have attempted to modify DNA repair pathways (Chu et al. 2015; Yu et al. 2015) to increase both the efficiency of HDR and the ratio of precise edits to imprecise mutations. These methods are most applicable for in vitro cell culture applications where potential toxicity is less limiting.
For short insertions, single-stranded oligodeoxynucleotide (ssODN) templates hold significant promise for treating disease because of their ease of synthesis. However, sequence changes encoded by the ssODN are infrequently incorporated after editing (<10 percent), and desired edits are typically outnumbered by other sequence outcomes (presumably from NHEJ). Strategies to link the ssODN to Cas9 help increase HDR (Carlson-Stevermer et al. 2017; Lee et al. 2017). Other methods avoid the use of HDR and leverage other DNA repair pathways (Suzuki et al. 2016).
Base editors are particularly attractive for clinical translation as they avoid DNA double-strand breaks entirely. They employ a catalytically dead version of Cas9 fused to a DNA deaminase to modify base pairs proximal to the sgRNA target, deaminating cytidine bases to form uridine. The modified bases are recognized by the cell as mismatched and corrected to thymidine (Komor et al. 2016). Current work in this area mostly focuses on cytosine (C) > thymine (T) (or analogous guanine (G) > adenine (A)) base conversions, although future versions will aim to allow modifications of any single base (Gaudelli et al. 2017). Additionally, a new strategy, called prime editing, also avoids DNA doublestrand breaks and has been demonstrated in vitro with mammalian cells for scarless incorporation of new sequences (Anzalone et al. 2019).
CHALLENGE 3: PRECISE TRANSCRIPTIONAL CONTROL
Even if challenges 1 and 2 are met with perfect accuracy and precision, expression of edited genes to generate RNA transcripts can vary over time as well as across cell differentiation and behavior patterns. Misregulation of the edited transcript can compromise therapeutic efficacy or lead to adverse events. Therefore, it is critical to consider strategies to maximize transcriptional control, especially when inserting new bases.
A striking discovery about the necessity of precise transgene expression recently emerged in the field of chimeric antigen receptor (CAR) T cell therapy. In the CAR T paradigm, a synthetic CAR transgene targeting a cancerenriched antigen is inserted ex vivo into a patient’s T cells, which are then expanded and reinfused, thereby engineering the immune system to recognize and target cells bearing the antigen (Piscopo et al. 2018). CRISPR-Cas9 was recently used to generate CAR T cells featuring a transgene at the T cell receptor alpha (TRAC) locus, which ensured that CAR expression was regulated by the endogenous TRAC promoter (Eyquem et al. 2017). These CAR T cells demonstrated promising results in a leukemic mouse model and also displayed fewer biomarkers of dysfunctional CAR T cells, suggesting that precise transgene control may yield a more potent cell product.
CHALLENGE 4: PRECISE EDITING IN SPECIFIC CELLS AND TISSUES
Precise delivery of editing components to the right cells and tissues remains a challenge as many delivery agents suffer from low efficiency, high toxicity, and immunogenicity, but viral and nonviral delivery agents have been engineered to achieve cell and tissue specificity.
Viral constructs can be engineered to harbor cell and tissuespecific promoters that drive expression of the gene editing system (Ran et al. 2015; Swiech et al. 2014) so that editing machinery is not expressed in nondesired cell types. And several nonviral designs have demonstrated high gene editing efficiencies when used with RNPs, ranging from 30 to 40 percent in cell lines, and up to 90 percent delivery efficiency (Chen et al. 2019; Mout et al. 2017; Sun et al. 2015; Zuris et al. 2015).
Custom biomaterials can also be engineered to direct genetic payloads to specific tissue types to allow gene editing in situ, thereby bypassing many of the biomanufacturing challenges associated with ex vivo cell therapy. Researchers recently developed DNA nanocarriers with the capacity to deliver CAR transgenes to T cells in a leukemic mouse model by coupling anti-CD3 ligands to polyglutamic acid (Smith et al. 2017). These nanocarriers demonstrated specificity to circulating T cells over other blood cell types shortly after delivery, causing tumor regression.
OUTLOOK
It is likely that strategies to meet these four challenges will be complementary, ultimately enabling more precise genomic surgery in patients’ cells. For in vitro applications, drug discovery will probably be accelerated by enhanced tools for disease modeling, target validation, and toxicological studies. For ex vivo uses, precision-engineered cell and tissue therapies may incorporate more functionality
from synthetic circuits (Weinberg et al. 2017). Finally, for in vivo somatic gene editing applications, injectable viral and nanoparticle strategies could specifically edit stem cells to regenerate tissues and correct disease-causing mutations.
Successful strategies to overcome the challenges described above may pave the way for a new wave of transformative therapeutics.
REFERENCES
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR. 2019. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature, doi:10.1038/s41586-019-1711-4.
Barrangou R. 2019. Thinking about CRISPR: The ethics of human genome editing. CRISPR Journal 2(5).
Carlson-Stevermer J, Abdeen AA, Kohlenberg L, Goedland M, Molugu K, Lou M, Saha K. 2017. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nature Communications 8:1711.
Chen Y, Liu X, Zhang Y, Wang H. 2016. A self-restricted CRISPR system to reduce off-target effects. Molecular Therapy 24(9):P1508–10.
Chen G, Abdeen AA, Wang, Y, Shahi PK, Robertson S, Xie R, Suzuki M, Pattnaik BR, Saha K, Gong S. 2019. A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nature Nanotechnology 14:974–80.
Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R. 2015. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nature Biotechnology 33:543–48.
Cradick TJ, Fine EJ, Antico CJ, Bao G. 2013. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Research 41(20):9584–92.
Cyranoski D. 2019. The CRISPR-baby scandal: What’s next for human gene-editing. Nature 566:440–42.
Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR. 2015. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nature Chemical Biology 11(5):316–18.
Duan J, Lu G, Xie Z, Lou M, Luo J, Guo L, Zhang Y. 2014. Genome-wide identification of CRISPR/Cas9 off-targets in human genome. Cell Research 24:1009–12.
Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJC, Hamieh M, Cunanan KM, Odak A, Gönen M, Sadelain M. 2017. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543(7643):113–17.
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. 2017. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551:464–71.
Hemphill J, Borchardt EK, Brown K, Asokan A, Deiters A. 2015. Optical control of CRISPR/Cas9 gene editing. Journal of the American Chemical Society 137(17):5642–45.
Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, Liu DR. 2018. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556:57–63.
Jasanoff S, Hurlbut JB, Saha K. 2019. Democratic governance of human germline genome editing. CRISPR Journal 2(5).
Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. 2016. Highfidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529(7587):490–95.
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. 2016. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–24.
Lander E, Baylis F, Zheng F, Charpentier E, Berg P, Bourgain C, Friedrich B, Joung JK, Li J, Liu D, and 8 others. Comment: Adopt a moratorium on heritable genome editing. Nature 567:165–68.
Lee K, Mackley VA, Rao A, Chong AT, Dewitt MA, Corn JE, Murthy N. 2017. Synthetically modified guide RNA and donor DNA are a versatile platform for CRISPR-Cas9 engineering. eLife 6:e25312.
Mout R, Ray M, Tonga GY, Lee Y-W, Tay T, Sasaki K, Rotello VM. 2017. Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for for efficient gene editing. ACS Nano 11(3):2452–58.
Mueller K, Carlson-Stevermer J, Saha K. 2018. Increasing the precision of gene editing in vitro, ex vivo, and in vivo. Current Opinion in Biomedical Engineering 7:83–90.
Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. 2013. High-throughput profiling of off-target DNA cleavage reveals RNAprogrammed Cas9 nuclease specificity. Nature Biotechnology 31(9):839–43.
Piscopo NJ, Mueller KP, Das A, Hematti P, Murphy WL, Palecek SP, Capitini CM, Saha K. 2018. Bioengineering solutions for manufacturing challenges in CAR T cells. Biotechnology 13(2).
Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154(6):1380–89.
Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, and 3 others. 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520(7546):186–91.
Ryder SP. 2018. #CRISPRbabies: Notes on a scandal. CRISPR Journal 1(6):355–57.
Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. 2016. Rationally engineered Cas9 nucleases with improved specificity. Science 351(6268):84–88.
Smith TT, Stephan SB, Moffett HF, Mcknight LE, Ji W, Reiman D, Bonagofski E, Wohlfahrt ME, Pillai SPS, Stephan MT. 2017. In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nature Nanotechnology 12:813–20.
Sun W, Ji W, Hall JM, Hu Q, Wang C, Beisel CL, Gu Z. 2015. Selfassembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angewandte Chemie 54(41):12029–33.
Suzuki K, Tsunekawa, Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, and 25 others. 2016. In vivo genome editing via CRISPR/Cas9 mediated homologyindependent targeted integration. Nature 540:144–49.
Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F. 2014. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nature Biotechnology 33(1):102–106.
Weinberg BH, Hang Pham NT, Caraballo LD, Lozanoski T, Engel A, Bhatia S, Wong WW. 2017. Largescale design of robust genetic circuits with multiple inputs and outputs for mammalian cells. Nature Biotechnology 35:453–62.
Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, Liu H, La Russa M, Xie M, Ding S, Qi LS. 2015. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16(2):142–47.
Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen Z-Y, Liu DR. 2015. Cationic lipidmediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nature Biotechnology 33:73–80.
This page intentionally left blank.








