2
Surveillance
The coronavirus disease 2019 (COVID-19) pandemic has exposed gaps in the capacity of worldwide national health systems and global-level systems to detect emerging and reemerging pathogens—including possible zoonotic threats, new strains of influenza with pandemic potential, and antigenic drifts in known viruses—before an outbreak occurs and a response is required. To close these gaps, countries need to collaborate to create early warning systems that are supported by political commitment, stable governance, and sustainable financing. Effective outbreak surveillance is urgently needed, since epidemics and pandemics are likely to become more frequent due to factors such as the expansion of urbanization, the growth and intensification of livestock production, more extensive and rapid global travel and trade connections, the effects of climate change, and pervasive socioeconomic inequities. Fortunately, the COVID-19 pandemic has also revealed the benefits of leveraging political will and financial resources to realize this early warning surveillance network for future emergent pathogens (Carroll et al., 2021).
PREPAREDNESS FOR SURVEILLANCE DURING THE COVID-19 PANDEMIC
Various indicators and indexes have been developed in recent years to evaluate countries’ level of preparedness, identify gaps and weaknesses, and support strengthening their capabilities to prevent, detect, and respond to outbreaks, epidemics, and pandemics of infectious diseases. For example, the Global Health Security Index (GHSI) draws on open-source informa-
tion to assess and benchmark health security and related capacities in 195 countries (JHU, 2019). Similarly, WHO’s Joint External Evaluations (JEEs) have been used for evaluating a country’s ability to prevent, detect, and respond to infectious diseases and outbreaks. Countries ranked higher in terms of preparedness according to GHSI, JEEs, and other indicators would be expected to respond more effectively to an actual pandemic event; however, that was not the case for COVID-19. An evaluation of the predictive value of GHSI and JEEs found that countries’ health preparedness scores were not correlated with detection response times or mortality outcomes (Haider et al., 2020). Furthermore, responsibilities for countries to act on their GHSI scores and improve preparedness are not necessarily delineated. A rank-based analysis of Organisation for Economic Co-operation and Development (OECD) countries’ ability to respond to COVID-19—based on total cases, deaths, tests, and recovery rates—found that their pre-pandemic GHSI preparedness scores did not predict their actual response; the scores tended to overestimate some countries’ preparedness and underestimate others (Abbey et al., 2020). A study evaluating the correlation between countries’ GHSI scores and measures of COVID-19 burden found no association between GHSI and rate of testing and, unexpectedly, a positive association between GHSI and cases and deaths (Aitken et al., 2020). An analysis of imported COVID-19 cases reported across 49 countries in sub-Saharan Africa (SSA) as of April 2020 found that the countries with high (1) GHSI scores, (2) likelihood of severe cases, and (3) government effectiveness rankings were not necessarily reporting a higher incidence of cases or more information per case. Such gaps in information could indicate undetected transmission and illustrate the difficulty of detecting and responding to asymptomatic cases (Skrip et al., 2021). More broadly, these disparities between predicted preparedness and actual response to a pandemic highlight shortcomings in the way preparedness has been assessed. Furthermore, given the increasing degree of global interconnectedness, “identifying and controlling spread of newly arising infectious agents is only as effective as the practices within the poorest performing countries” (Aitken et al., 2020, p. 354).
ROLE OF SURVEILLANCE IN MITIGATING RESPIRATORY VIRUS OUTBREAKS
Surveillance has different functional roles in the context of a respiratory virus outbreak: (1) detecting potential new threats outside of a jurisdiction that could potentially be imported and spread locally, including epizootic, zoonotic, and epidemic threats; (2) detecting the importation and community transmission of an identified outbreak threat in animals and humans; and (3) assessing the extent and severity of an outbreak using forecasts and models. The first two roles focus on detecting a threat quickly and accurately, while
also minimizing false-positive test results; in some scenarios, these efforts may warrant oversampling high-risk locations (e.g., ports of entry). In contrast, activities to fulfill the third role—quantifying the spread—are situated in the realm of systems-based processing of large volumes of human samples, collected in a representative way, to understand the current number of cases and how that epidemiological landscape is changing.
Syndromic Surveillance for Infectious Diseases
Over the past two decades, syndromic surveillance has been used as a strategy for detecting and monitoring public health events based on individual- or population-level indicators in advance of confirmed diagnoses of an emerging infectious disease. For example, data on symptoms or clinical diagnoses such as influenza-like illness (ILI) or severe acute respiratory illness could serve as early indicators that an unusual respiratory pathogen is circulating (van den Wijngaard et al., 2008). These indicators and data on pneumonia of unknown origin were used retrospectively by Chinese authorities and WHO to assess evidence for early cases of COVID-19 (WHO, 2021b). Syndromic surveillance is theorized as advantageous for early detection of infectious disease outbreaks, given the time lags between initial symptoms and a clinically or laboratory-confirmed diagnosis (Chu et al., 2012). A retrospective study found that syndromic data from health registries—including on work absenteeism, general practice consultations, prescription medications dispensed, diagnostic test requests, hospital diagnoses, and deaths—correspond to patterns in respiratory pathogen activity and thus can be used for surveillance (van den Wijngaard et al., 2008). Beyond the early detection value, syndromic surveillance data can also inform public health actions, contribute to improved situational awareness, and bolster the credibility of public communications. Clinical laboratory testing that includes signs and symptoms, with data aggregated in the cloud, can also serve as a type of surveillance when syndromic trends are reported, including negative test results (Meyers et al., 2018). If enough negative results are reported in a certain region linked to people with severe symptoms, it could be a signal of a new pathogen and trigger the need for additional testing. This was used during H1N1 in 2009 and again in Wuhan when the outbreak first began; syndromic panels were negative for pneumonia, leading to identifying a novel virus. Incorporating health systems, including academic health institutions and the data they collect, can strengthen global public health infrastructure, even for pathogens not often targeted by surveillance. This is an opportunity to add scale and capacity to public health.
Box 2-1 describes some of the major influenza surveillance collaboratives that serve to do so. Syndromic surveillance conducted alone, such as with ILI systems, or in combination with viral testing can be and has been used to
track activity in real time during a pandemic (Brammer et al., 2011; Lipsitch et al., 2009; Shaman et al., 2011). However, a qualitative study of syndromic surveillance during the 2009 H1N1 pandemic in Ontario, Canada, found that it had only a limited impact on decision making about public health response activities, which were largely informed by logistics (e.g., vaccine availability) and traditional forms of surveillance using laboratory data (Chu et al., 2012).
ONE HEALTH APPROACH
The existing wealth of knowledge and evidence-based strategies for mitigating epidemic and pandemic threats remains largely untapped and underused. For instance, “One Health” is a collaborative, multilevel, transdisciplinary approach that aims to achieve optimal health outcomes between people, plants, and animals in their shared environment (CDC, n.d.). It is increasingly recognized by governments, scientists, the private sector, nongovernmental organizations, academic partners, and others as an effective way to combat health threats that affect people, animals, plants, and the shared environment. One Health approaches are particularly relevant to emerging infectious diseases, of which greater than 60 percent are zoonotic, and to diseases that have a strong link to environmental conditions (e.g., water- and vector-borne diseases). Many of these diseases spill over to humans through a complex and multi-step process (see Figure 2-1). This type of surveillance effort is especially critical in low- and middle-income countries (LMICs) and places that have an extensive human–animal interface.
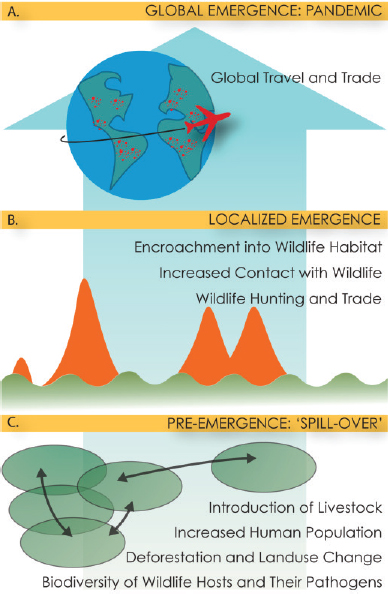
SOURCE: Bogich et al., 2012.
In one review of nearly 400 public health events of international concern, a breakdown or absence of public health infrastructure was identified as the driving factor for just under 40 percent of outbreaks. Though many outbreaks do not result in global pandemics, pandemic prevention at the local level should include stronger public health infrastructure, expanded surveillance, and incorporation of development agencies into strategies that target where populations intersect with the environment (Bogich et al., 2012).
Evidence for One Health Approaches and the Need for This Type of Surveillance
Earlier studies have recommended active surveillance through One Health approaches to mitigate infectious disease threats as discussed during an Institute of Medicine workshop on emerging viral threats (IOM, 2015). Some of these recommendations have been implemented, but not to the extent that outbreaks such as COVID-19 could have been prevented, despite the warning signs and knowledge about how to use One Health approaches to intervene. For example, forming One Health outbreak investigation teams that involve veterinarians, medics, social scientists, wildlife biologists, and ecologists could enable more rapid investigation of the zoonotic origins of emerging diseases, something that was not a focus of early COVID-19 investigations. Done well, One Health approaches can lead to higher returns on investment through joint human–animal disease surveillance and control measures (Kelly et al., 2020). Ongoing exercises working across sectors also help to facilitate collaboration and connect stakeholders that do not typically interact, improving future communications.
Interagency One Health platforms have been launched in LMICs specifically to link operations of ministries of health, agriculture, and the environment and wildlife, while also maximizing surveillance for influenza and other emerging zoonoses. Because South Asia has been identified as a “hot spot” for emerging zoonotic disease, it has focused on strengthening One Health efforts since the early 2000s, along with many bilateral and multilateral partners. For example, Bangladesh has seven One Health research programs, offers three One Health postgraduate degrees at various universities, and has field epidemiology training programs for public health and laboratory personnel through the U.S. CDC (McKenzie et al., 2016). It also developed a Strategic Framework for One Health Approach to Infectious Diseases in 2012, which was endorsed by the Ministry of Health with widespread support (IEDCR, 2012).
Through multi-sector collaboration in Kenya, the government developed an institutional framework to highlight the importance of several types of diseases that informs capacity building programs, surveillance, and workforce development, among other areas. It has noted improved
outbreak response and newly generated data that informed disease control programs and increased preparedness (Munyua et al., 2019). The ability to more easily share disease outbreak information across sectors and rapid response at the county level has been credited with reducing spillover to humans in an anthrax outbreak in 2016. More recently, the country has deployed a syndromic surveillance system in domestic and wild animals, using a mobile phone application for reporting and analysis, with hopes to improve real-time surveillance within the animal health sector. Stakeholders involved in this effort believe that “the adoption of the One Health program and approach in Kenya has led to rapid detection and control of zoonotic disease outbreaks at their source and thereby enhanced global health security” (Munyua et al., 2019).
Lessons from COVID-19
The emergence of SARS-CoV-2 has reinforced the rising risk of pathogens that are capable of jumping species to humans. Despite a wealth of evidence on different wildlife species and the types of viruses they carry, the connection between that knowledge and what measures are needed to reduce the risk of spillover is more tenuous. After SARS in 2003, substantial research in China and Southeast Asia demonstrated a wide diversity of related viruses in wildlife (bats in particular) and that some of these were able to infect human cells in vitro and cause SARS-like disease in mice with human ACE2 receptors (Ge et al., 2013; Latinne et al., 2020; Menachery et al., 2015). Farming wildlife known to act as SARS intermediate hosts continued to expand, with around 14 million people employed in the industry in China alone in 2016 (UNDP China, 2017). Furthermore, published evidence revealed that people in rural China were infected by bat SARS-related coronaviruses even without direct involvement in hunting or consuming wildlife (Wang et al., 2018). These studies were widely cited in the literature and cited by WHO in the rationale to include SARS-related coronaviruses as “priority pathogens” for vaccine and therapeutic development through the R&D Blueprint effort. A small number of researchers globally were funded to develop therapeutics (e.g., remdesivir). However, efforts to close down wildlife farms, markets, and trade networks or remove known coronavirus hosts were not widely undertaken until after the COVID-19 outbreak began. Likewise, widespread funding of vaccine or therapeutic development through the U.S. National Institutes of Health, Coalition for Epidemic Preparedness Innovation, or R&D Blueprint did not occur prior to the outbreak. This example demonstrates that the knowledge generated through One Health approaches and research can provide important insight, but until the political motivation exists to act on the findings and provide funding, the problems will remain unsolved and likely surface again.
After SARS in 2003, China instituted a program of syndromic surveillance that did form part of the early warning system for clusters of pneumonia cases that were later diagnosed as COVID-19. The improved surveillance and laboratory capacity in 2020 was able to recognize the novel outbreak within just a few weeks of syndromic surveillance clusters (Chan et al., 2020).
Challenges to Achieving an Effective One Health Approach
Many experts recognize the barriers to widespread, practical implementation of a One Health approach across contexts. Ultimately, inadequate funding mechanisms, lack of incentives for collaboration, and competing interests across different government ministries pose major barriers to implementing One Health principles. Although some countries have created dedicated One Health task forces and crosscutting mechanisms linking ministries of health, environment, agriculture, and wildlife, they have been ineffective at scale and at the global level. Additionally, a review of One Health literature in 2017 found few efforts to systematize metrics and truly evaluate outcomes versus merely modeling projections (Baum et al., 2017). Of more than 1,800 papers, only 7 reported quantitative outcomes, and even these did not use a shared methodology. Without a standardized framework to capture metrics for these types of approaches, it will be difficult to encourage more widespread adoption of One Health. Another concern highlighted by multiple sources is sustainability of programming. A collection of three case studies in Africa concluded that broad institutional changes and sufficient funding are needed for One Health to become a more common approach to health policy at the national levels, and each country will need its own individualized plan based on its needs and capacities (Okello, 2014).
A key challenge that has been highlighted by the COVID-19 pandemic is a lack of connection between the evidence of a potential pandemic threat and forming policies to deal with it. Evidence that viruses related to SARS-CoV were present in wildlife and livestock in China was funded by research agencies and published in scientific papers but not brought into a formal risk assessment framework. Similarly, wildlife farming and trade were considered the likely causes of the emergence of SARS, but policies to conduct coronavirus surveillance as a routine for wildlife hunters, farmers, or traders, or the animals they sold were not formalized into the public health system. Collecting influenza samples and identifying strains from wild birds and farmed animals is routine in some countries but could be expanded in many others. However, challenges include difficulties in assessing the pandemic potential of novel strains, as sequencing and pursuing all of those identified would quickly exhaust available resources and workforce.
To be optimally effective, One Health collaborative approaches should be truly international—not just interagency—and leverage the power of established regional and global health organizations. Expanded collaboration among national development agencies, such as the United States Agency for International Development, as well as multilateral organizations (e.g., WHO, Food and Agriculture Organization of the United Nations, World Organisation for Animal Health, United Nations Environment Programme, and World Bank and regional equivalents), should also be encouraged in the One Health sphere. More areas for additional research to inform this approach can be found in Box 2-2.
RELEVANT FINDINGS AND CASE STUDIES FROM THE COVID-19 PANDEMIC
This section provides an overview of COVID-19 findings and case studies that demonstrate successes, highlight innovations, and illustrate challenges related to surveillance for respiratory pathogens.
Core Public Health Functions for Surveillance
Core public health functions for surveillance include identification and notification, sampling and genomics, and testing and contact tracing for event notification and control. Strengthening these capacities will be critical to more effectively prepare for and respond to future epidemic and pandemic events.
Tracking Outbreak Progress
Pandemic statistics—particularly the proportion of the population infected—are believed to be generally and substantially underestimated. Contributing factors related to the limitations of classical surveillance approaches include insufficient diagnostic capacity, failure to detect asymptomatic cases rapidly enough, and political shortcomings of following through on outbreak predictions. The COVID-19 pandemic has included cases in which traditional surveillance methods have underestimated the actual prevalence. For example, polymerase chain reaction (PCR) testing for detecting infections is hampered by limited testing capacity, high rates of false-negative results, and the test’s inability to detect asymptomatic and subclinical infections (Silverman et al., 2020). An analysis of the use of influenza surveillance networks to estimate U.S.-state-specific SARS-CoV-2 prevalence has suggested that during the early phases of the pandemic, greater than 80 percent of infections were undetected (Silverman et al., 2020). Hospital-based surveillance has limited utility in accurately estimating the number of cases, because many people who test positive are not hospitalized (Alwan, 2020), or there are delays in obtaining timely clinical data (Garg et al., 2020). However, in some localities, it may have contributed to mitigating the initial spread. For instance, in Singapore, such a surveillance and containment strategy has been documented as contributing to improved case ascertainment and slowing transmission (Ng et al., 2020). Telehealth data could also contribute to surveillance systems in a pandemic context, particularly if many patients are not hospitalized and virtual visits are encouraged as an infection control measure (Koonin et al., 2020).
Sampling and Genotyping
Laboratory science is a cornerstone of successfully controlling an epidemic or pandemic. Core components of an effective laboratory response include (1) building testing capacity early, (2) preparing the workforce for a dynamic response, (3) strengthening information management systems, and (4) creating laboratory partnerships that can be leveraged during an event (McLaughlin et al., 2021).
During the COVID-19 pandemic, genotyping and genomic surveillance have been valuable tools for detecting new variants and understanding their potential effect on infectivity and health outcomes (CDC, 2021a). Understanding the genomic diversity of an infectious pathogen can inform more effective strategies to contain its spread during the initial stages of an outbreak. For example, in the highly interconnected region spanning Maryland and Washington, DC, in the United States, more than 2,500 cases of COVID-19 were reported within 3 weeks of the first detected case in
March 2020. Genomic sequencing analysis of 114 complete viral genomes revealed a broad diversity that included all the lineages that were known to be circulating globally at the time, signifying that multiple introductions of the virus into the region were likely. Moreover, a combined analysis of those genomes with clinical metadata determined that clinically severe cases had originated from all the major lineage strains (Thielen et al., 2021). Genomic sequencing is also valuable for seasonal and pandemic influenza (CDC, 2021b).
However, this level of genomic surveillance is not universally conducted, making it difficult to obtain a global view of dominant strains in different areas. While SARS-CoV-2 led to an acceleration of efforts, without supplemental epidemiology and surveillance data, the genomic sequencing is not sufficient to show which strains are more transmissible or more lethal (Morgan et al., 2021). Some countries cannot afford the technology, nor do they have sufficient workforce; others, such as the United States, have not invested in the infrastructure because it was not seen as widely important until recently and fragmented data systems make it difficult to coordinate and share across institutions. For example, the United States was ranked thirty-third in the world during this pandemic, with less than 2 percent of cases sequenced. The COVID-19 Genomics UK Consortium (COG-UK), set up in April 2020, is an example of what a well-functioning system would look like and that others could model.1 After just 1 year, COG-UK had sequenced more than 450,000 genomes, contributing to the United Kingdom’s rank of fifth in the world, sequencing more than 8 percent of its cases (Maxmen, 2021). COG-UK has a long-term goal of developing a sustainable sequencing network across the United Kingdom. The consortium includes partners from the National Health Service, public health agencies, academic partners, lighthouse labs, and the Wellcome Sanger Institute.
Individual- and Population-Level Testing
At the individual level, COVID-19 testing strategies include quantitative PCR (qPCR), loop-mediated isothermal amplification, and antigen testing performed at the point of care, central laboratories, or through rapid testing modalities. At the population level, testing strategies range from pooled testing to screening to surveillance of wastewater and surfaces. However, the optimal strategy for a given setting is context specific and not necessarily universal—different approaches are warranted to serve various purposes (Mina and Andersen, 2021). Diagnostic testing aims to identify people with SARS-CoV-2 infection, for both clinical management and isolation, contact
___________________
1 For more on the COVID-19 Genomics UK Consortium, see https://www.cogconsortium.uk/cog-uk/about-us (accessed August 28, 2021).
tracing, and contact testing. Although the relatively lengthy time to results from laboratory-based PCR testing reduces its utility in preventing transmission, rapid point-of-care tests can enable more widespread testing coverage. The goals of surveillance testing are to conduct representative sampling to estimate prevalence and inform response activities at the population level. Antibody testing can be used to understand the breadth of historical exposure, while ongoing community transmission can be monitored through PCR testing of wastewater or pooled testing in low-prevalence settings, for example. Screening, which includes entry screening and public health mass screening efforts, can be used to detect people who are a- or pauci-symptomatic but may be infectious. Rapid antigen tests can offer reduced costs and short turnaround times, particularly in places where reverse transcription (RT)-PCR capacity is limited. For COVID-19 screening, rapid antigen tests perform best in presymptomatic and early symptomatic cases with high viral load up to 5 days from symptom onset. Their shorter turnaround times for results and lower costs can facilitate community testing regardless of symptoms in homes, care settings, and workplaces that may face risks of high levels of community transmission (ECDC, 2021).
The COVID-19 pandemic has demonstrated the extent to which virological, genotyping, and population-wide serological surveillance can be limited by testing capacity (de Lusignan et al., 2020). Efforts to bolster preparedness for future events are ongoing in countries in Africa and other regions to strengthen testing and other capacities to enhance public health surveillance systems, such as by integrating pathogen genomics (Inzaule et al., 2021). Evidence suggests that programs with expanded testing capacity during COVID-19 effectively curtailed transmission. In late 2020, Slovakia implemented a strategy of population-wide rapid antigen testing and additional restrictions on social contact in 45 counties; modeling suggests that these measures—as well as isolation of household contacts—were associated with a 58 percent reduction prevalence within 1 week of implementation (Pavelka et al., 2021).
Contact Tracing
Contact tracing can contribute to not only curbing transmission of an infectious disease threat—via identifying and isolating exposed contacts—but also reducing case fatality rates through early detection and referral to care (Yalaman et al., 2021). Drawing on evidence from 138 countries, an analysis of different contact tracing strategies and COVID-19 case fatality rates found that comprehensive contact tracing—along with appropriate case isolation—was associated with significantly reduced case fatality rates, even after controlling for public health and social measures and the number of tests performed (Yalaman et al., 2021).
Contact tracing was also demonstrated to be successful during the West African Ebola outbreak in 2014–2015, most notably in Nigeria. Thanks to establishing a field epidemiology and laboratory training program in 2008, hundreds of Nigerian health workers were trained in contact tracing, outbreak investigations, and report development. The Nigerian program was the first to apply the concept of One Health in full, because it included epidemiology, laboratory, and veterinary tracks (Oleribe et al., 2015). The teams had worked together through Lassa Fever and polio outbreaks and were designed to be interdisciplinary, so they were poised to respond rapidly to the Ebola outbreak and can be a model for future outbreaks.
During the COVID-19 pandemic, few countries had a sufficient number of trained personnel to conduct contact tracing, particularly during the early stages. Even settings where contact tracing was initially effective faced difficulties in sustaining those efforts as the pandemic unfolded. In Germany, initial contact tracing was largely successful until capacity became overwhelmed as infections peaked (Loh, 2020; Reintjes, 2020). In summer 2020, as case numbers began to decline, many countries devoted more resources to increasing testing capacity than to building contact tracing capacity or ensuring that people who became infected could appropriately isolate (Loh, 2020).
A review of COVID-19 contact tracing efforts in Nigeria, Rwanda, South Africa, and Uganda provided several best practices, challenges, and lessons informing future implementation of these efforts. The common challenges identified across all five countries include internal stigma, community resistance, and apathy driven by mistrust and perceived and internal stigma. Another critical common challenge was misinformation and an overwhelming load of contact tracing and case detection workload for health care workers. For example, the number of contact tracers per 100,000 population ranged from a low of 3 in some areas to a high of 186. Other challenges identified included fears around contact tracers’ risk of COVID-19 infection, limited testing and health care capacity, mistrust of political entities, and poor adherence to quarantine and isolation guidelines and rules. Lessons learned from these nations broadly included the effectiveness of decentralizing and building capacity for communication, contact tracing, testing, and their associated human resources at the local and community levels. Additionally, the authors found that interoperable data and technology should complement traditional contact tracing efforts to improve decision making. Partnerships, meaningful community engagement, and coherent political leadership were identified as mechanisms to build trust, combat misinformation, and scale interventions (Nachega et al., 2021).
The general effectiveness of contact tracing varies across settings and contexts. An analysis of COVID-19 countermeasures in Yamagata Pre-
fecture, Japan, found that retrospective contact tracing efforts to identify epidemiological links were likely effective in halting the first wave (January–May 2020) (Seto et al., 2021). According to a mathematical modeling study, a combination of highly effective contact tracing and isolation was sufficient to bring a new outbreak under control within 3 months, but the likelihood of control declines if fewer cases are detected through contact tracing (Hellewell et al., 2020). A simulation study found that a testing capacity of 0.7–9.1 tests per 1,000 population would be needed to contain the spread of SARS-CoV-2, depending on public health and social measures in place, and that the number of new daily infections did not always decline—it could exponentially increase if contact tracing and testing efficacy fell lower than 60 percent (Fiore et al., 2021).
Variations in the effectiveness of contact tracing can be attributable to the number of observed asymptomatic infections, transmission efficiency, population distribution and size, and the size of the secondary infection cluster. This suggests that when developing testing and contact tracing strategies, policy makers should consider population-level density, geographical distribution, and travel behavior (Fiore et al., 2021). Moreover, most strategies mainly employ a “forward-tracing” protocol to notify people that they were exposed to a known case. However, a bidirectional tracing approach also includes reverse tracing, which seeks to identify the parent case who infected the known case, as well as other cases related to that parent case. A modeling study has suggested that bidirectional tracing is a more robust approach to outbreak control for COVID-19, yielding a reduction in the effective reproduction number (Rt) more than twofold greater than forward tracing alone (Bradshaw et al., 2021).
CHALLENGES IN PUBLIC HEALTH SURVEILLANCE HIGHLIGHTED DURING THE COVID-19 PANDEMIC
COVID-19 has revealed multiple limitations of current public health surveillance systems and tools, which were primarily designed for ongoing surveillance of known pandemics and seasonal influenza rather than for early detection and mitigation of respiratory pathogens with pandemic potential. These existing systems are also unable to accommodate the sustained surge capacity necessitated by a large-scale global pandemic event. Specific challenges that undermine the ability to conduct syndromic surveillance and interpret surveillance data include (1) the effect of media reporting early on; (2) changes in health-seeking behavior driven by pandemic-control measures, such as physical distancing; and (3) changes in systems for clinical coding and patient management (Elliot et al., 2020). Strengthening central systems for data use, collection, and sharing would allow for more effective quantification of the spread of infection and optimal
parameterizing for forecasts and models. Furthermore, in addition to the strategies described below, core surveillance capacities can be strengthened through reiterative testing via simulations and other exercises (Reddin et al., 2021).
Ascertainment Bias
Ascertainment bias is a consequence of biased sampling that has likely undermined efforts to estimate the burden, infectiousness, and fatality of SARS-CoV-2 since the outset of the pandemic (see Box 2-3). Such biases can misinform the public about the severity of a disease and the utility of public health interventions in general or for particular subgroups (Peixoto et al., 2020). For example, using case fatality rates based on hospital fatality rates, which include only a small subset of individuals with the disease, has led to misunderstanding that alarmed the public and inaccurate comparisons of disease severity between COVID-19 and Ebola (Winters et al., 2020).
Inaccurate epidemiological estimates based on nonrepresentative or inaccurate data have also led to ill-advised policy decisions and ineffective responses. For months at the outset of the pandemic, the U.S. government was unable to estimate how many people were sick with COVID-19, were hospitalized, or had died (Meyer and Madrigal, 2021). Levels of com-
munity and background transmission were underestimated in many U.S. localities, which could have been rectified by routine standardized testing, which can mitigate sampling bias (Angelopoulos et al., 2021). Instead, the White House Coronavirus Task Force relied on forecasts from the Institute for Health Metrics and Evaluation during the initial days of the pandemic (IHME, 2020). This model made a number of inaccurate assumptions: that the epidemic curve would follow the outbreaks in China and Italy (Holmdahl and Buckee, 2020), that physical distancing measures would remain effective, and that social distancing was being implemented the same everywhere (Jewell et al., 2020). Its initial projection—a death toll of only around 60,000 U.S. individuals (which was actually surpassed by May 2020)—influenced state and federal officials to pivot to reopening the economy instead of prolonging physical distancing and other public health measures (Cancryn, 2020). Other countries, such as Brazil, reduced testing, resulting in underreporting disease incidence (Fonseca, 2021). In contrast, China demonstrated the value of improved testing strategies. Early in the outbreak, diagnosis was based only on testing that had severe capacity constraints; when the case definition was expanded to include radiological criteria as an adjunct, it contributed to elucidating the true infection rates (Tsang et al., 2020). Additional research needs related to ascertainment biases can be found in Box 2-4.
Variability in Estimating Infectiousness and Fatality Rates
Wide variability has been observed between country- and state-level COVID-19 infectiousness and fatality rates. For instance, a Bayesian modeling study—which was designed to minimize ascertainment bias—analyzed confirmed data on COVID-19 cases, deaths, and recoveries from U.S. states
and countries in all world regions. By April 2020, estimates of infectivity ranged from 9–13 unreported cases for every confirmed case globally. At the outset of the pandemic, the estimated mean global reproduction number and total infection fatality rate were 3.3 (confidence interval [CI] 1.5, 8.3) and 0.17 percent (CI 0.05–0.9 percent), respectively. By mid-April, estimates for those values had evolved to 1.2 (CI 0.6, 2.5) and 0.8 (CI 0.2–4.0) percent. Moreover, the variability observed between the country- and state-level values implies substantial uncertainty about the ability to accurately assess an epidemic’s current state or trajectory (Chow et al., 2020). In terms of the earliest fatality risk estimates, a crude case fatality rate of 3.67 percent was found among cases from mainland China (Verity et al., 2020) and a case fatality risk of 1.4 percent in Wuhan (Wu et al., 2020). In contrast, an early analysis in Italy found crude case fatality rates of 10.6 percent nationwide and 18.3 percent in Lombardy, much higher than the rates estimated based on data from outbreaks in China and aboard the Diamond Princess cruise ship (1 percent) (Vicentini et al., 2020).
Clinical Icebergs
COVID-19 exemplifies challenges caused by the “clinical iceberg” phenomenon: the relative proportions of clinically observed infections versus unobserved infections. Quantifying those proportions is critical to developing the parameters for models to elucidate population-level transmission dynamics and epidemic trajectories that are needed to inform public health policy (Wu et al., 2020). Most cases of respiratory pathogens in particular go undiagnosed and not notified, underscoring the need for surveillance and detection systems that “search for the unexpected” and unseen.
Underrepresentative Sampling Frames
A general limitation of survey-type seroprevalence studies is the under-representation of vulnerable populations at high risk of COVID-19 infection and/or mortality (e.g., residents of nursing homes, persons who are homeless, persons who are incarcerated, or persons living in large urban slums or refugee camps) due to challenges related to reaching and sampling those populations. Lack of representativeness in sampling frames undermines the ability to accurately estimate COVID-19 prevalence (Bendavid et al., 2021) and SARS-CoV-2 seroprevalence, as exemplified in a study that inferred the total number of people infected in all of Croatia from a serosurvey of just two factories (Ioannidis, 2021). However, some approaches have achieved a much greater degree of sampling representativeness, thus yielding more accurate estimates. Luxembourg implemented a mass screening program during the early summer 2020 wave of COVID-19 that aimed to capture
a representative sample of the entire population—including residents and cross-border workers—and found a significant attack rate among asymptomatic cases (Wilmes et al., 2021). Similarly, a representative, nationwide, population-based serological survey in Spain reported that at least one-third of people with COVID-19 were asymptomatic, underscoring the importance of early testing and detection (Pollán et al., 2020). Related, although it was against the convention early in the pandemic to only test individuals with a connection to China, the Seattle Flu Study was one of the first to find community transmission in the United States by testing study samples for SARS-CoV-2 (Chu et al., 2020). This further illustrates how diagnostic testing can be an effective response, that each community will have different levels of risk, and testing may be more useful in some circumstances more than others (Sharfstein et al., 2020). Further needs for research related to transmission are highlighted in Box 2-5.
Strengthening Reporting
Institutional hierarchies and bureaucracies can stifle reporting on the progress of an outbreak. For instance, officials may be hesitant to trigger investigations by reporting on diseases of concern due to fear of stigma or the economic implications, as observed with Ebola in central and West Africa. Other layers of bureaucracy in reporting can also delay a response. For instance, within the African integrated disease surveillance response system, health facilities report to district and national levels of health authorities on priority diseases of importance; these then report to global institutions, which can be slow to respond. Reporting could also be strengthened by involving the communities—training and providing tools to frontline public health workers and community workers in accurately detecting, reporting, and analyIng during routine public health surveillance for priority diseases; this can help to ensure timely, complete, and accurate data for decision making. Using a case study designed to train resident epidemiologists in
Liberia, it is possible for 1–2 instructors to train up to 20 students in collecting useful data, auditing data quality, and conducting Strengths, Weaknesses, Opportunities, Threats analyses (Frimpong et al., 2017). Incentives are another mechanism to strengthen reporting during outbreaks. Different incentives may be most effective at different levels, such as country-level incentives of funding and resources, increased prestige for effective systems, and international encouragement (IOM and NRC, 2009).
Zoonotic Surveillance
The interface between humans, domesticated animals, and wild animals is a major locus for the emergence of zoonotic diseases, which comprise the majority of emerging infectious disease threats reported worldwide. Coronaviruses and avian influenza viruses are among the foremost zoonotic threats to human health (Huong et al., 2020), with evidence suggesting that SARS-CoV-2 may be due to zoonotic transmission that may have originated in or been subsequently amplified at a live animal market in Wuhan (Tiwari et al., 2020; WHO, 2021b). The COVID-19 pandemic and other epidemics of zoonotic origin of recent decades have underscored the need for One Health approaches to strengthen zoonotic surveillance efforts, detect viral strains with larger antigenic drifts, and develop better strategies to understand the degree of potential threat posed by emerging strains.
Live Animal Markets
Live animal markets provide ideal conditions for zoonotic transmission through an intimate interface among animals and people, leading to amplification of pathogen load and the potential for recombination or selection pressure for evolution of virulence. These factors highlight the need for more effective surveillance strategies in these settings (Tiwari et al., 2020). Multiple zoonotic influenza viruses have been associated with human exposure to animals at these types of markets and further down wildlife and poultry supply chains. An evaluation of wildlife supply chains for human consumption in Vietnam (2013–2014) used PCR testing to detect coronavirus sequences, finding high proportions of positive samples in field rats (34.0 percent) to be consumed by humans and among bats in guano farms near human residences (74.8 percent) (Huong et al., 2020). The analysis also found a mix of different types of bat and avian coronaviruses in rodent feces, suggesting that the mixture and amplification of coronaviruses along the wildlife supply chain to retail and restaurant settings could increase the potential for zoonotic spillover to consumers.
During the epidemic of avian influenza A H7N9 (2013–2015), which causes human infections primarily via zoonotic transmission, closing live
poultry markets in mainland China temporarily halted outbreaks (Peiris et al., 2016). However, such measures are not feasible over the long term, due to the country’s existing systems for live poultry production and marketing. In China and other countries in Asia, live poultry systems dominate poultry consumption. These systems, which are complex and do not tend to be intensively regulated, span a large network of farm production, transportation to wholesale markets, and retail distribution (Peiris et al., 2016). More sustainable and less disruptive approaches to reducing the risk of emergence and transmission of zoonotic influenza include instituting market “rest days,” banning live poultry in markets overnight, and separating terrestrial poultry from live ducks and geese; such strategies have been progressively implemented in Hong Kong (Peiris et al., 2016). Alternative strategies for reducing the risk of zoonotic viruses, beyond simply banning all live animal markets, have concomitant environmental and social benefits, including encouraging smaller-scale meat production, improving market hygiene, implementing more stringent regulations at markets, and outlawing the trade of certain wildlife (Petrikova et al., 2020). Others have argued that banning wildlife trade would effectively bolster the black market, so tighter regulation would be more effective (Tiwari et al., 2020).
Coordination and Assessment
An analysis of WHO’s JEE reports looked at trends in preparedness for high-consequence zoonotic infectious diseases among SSA countries (Elton et al., 2021). The veterinary workforce had the highest average score in all categories across all countries evaluated; response mechanisms had the lowest average score. Most countries provide public health training courses for veterinarians. The Southern African region had the highest mean score for all zoonotic disease categories. All five of the most frequently cited zoonoses on priority pathogen lists in SSA were neglected diseases: rabies, highly pathogenic avian influenza, anthrax, brucellosis, and bovine tuberculosis (TB). These findings suggest that SSA countries should leverage the convergence of public health, veterinary, and environmental government departments across African and global health organizations—such as the One Health consortia and the Pan-African network PANDORA-ID-Net—to implement a collaborative One Health approach to pandemic preparedness and response (Elton et al., 2021).
With increased genomic capacity for detection of novel viruses, a better strategy to assess risk of novel agents is needed. The recently published SpillOver is a comprehensive, publicly accessible risk assessment tool for systematically evaluating novel infectious viruses’ potential for zoonotic spillover and spread. Although data gaps limit the ability of SpillOver to rank relative animal–human transmission risk, among other challenges,
the tool and its associated watch lists can support virus discovery efforts to detect new animal viruses, assess and communicate risk, and inform pandemic preparedness and response efforts (Grange et al., 2021). It also offers opportunities for global collaborative research to understand the biology of pathogens that may be emergent and screen therapeutic agents in advance. Ideally, this would be available as a global repository of information that can be accessed widely by researchers.
Data Collection, Use, and Sharing
Digitally enabled public health strategies augmented by data science can aid in population surveillance, case identification, contact tracing, and evaluation of interventions based on mobility data and public communications. Harnessing the power of digital technologies through a combination of mobile phones, large online datasets, interconnected devices, low-cost computing resources, machine learning, and natural language processing underpins these efforts. Efforts are now focusing on ways to effectively and ethically incorporate data from digital and Web-based sources into public health surveillance—for example, through hybrid approaches that integrate data from traditional sources with data collected from Internet search queries, posts on social media networks, and other forms of open-source and crowdsourced data (Aiello et al., 2020). Metadata and line lists can be one informative way of linking data streams (Xu et al., 2020). In a pandemic, social media can serve as a powerful mechanism for communicating and disseminating information. However, a scoping review found that social media data were not leveraged for real-time surveillance to detect or predict cases during the COVID-19 pandemic as they have been for other infectious diseases, such as malaria and influenza (Tsao et al., 2021).
Within nations, sharing surveillance data across communities can be vital for identifying an outbreak’s impact (Liverani et al., 2018). However, two-way accountability is needed for entities with capacity to take immediate action on data and surveillance information that is shared by countries. Inefficiencies in collecting and sharing data among health agencies and across countries impeded the flow of information on critical treatment, patient, and event-level data during COVID-19 (Cossgriff et al., 2020). Another critical need is to standardize and harmonize data to enable data sharing (Fukushima et al., 2018). Additionally, practical issues, such as the location and method of long-term storage and maintenance, access control, and funding, are unresolved for epidemiologists and public health researchers (Pisani and AbouZahr 2010). Nevertheless, a model that has been used for event notification is Participatory One Health Digital Disease Detection; community members use smartphone and other web applications to report unusual disease events in humans and both wild and backyard animals;
these reports lead to a local response from health experts (Ending Pandemics, 2021). Another possible model to incentivize data reporting and sharing is the Global Initiative on Sharing Avian Influenza Data, which fosters collaboration by requiring data users to provide credit to submitters and also work to include them in joint viral data analyses (LoTempio et al., 2020).
Digital Contact Tracing Technologies
Functions of digital contact tracing technologies include outbreak response, proximity tracing, and symptom tracking (Anglemyer et al., 2020). During the COVID-19 pandemic, digital contact tracing has successfully complemented traditional tracing methods by using smartphone application technology to identify exposed social contacts, particularly if they are strangers (Rodríguez et al., 2021). To augment traditional approaches, countries such as South Korea, China, and Singapore have implemented digital contact tracing strategies that are regarded as having contributed to successfully controlling spread (Lancet Digital Health, 2020). Modeling studies suggest that digital contact tracing can break chains of transmission (Salathé et al., 2020), but robust evidence for its effectiveness in real-world outbreak settings is currently lacking (Anglemyer et al., 2020). Widespread implementation of this approach has been hindered by poor integration of the technology with existing surveillance tools (Anglemyer et al., 2020) and by ethical and legal concerns, particularly around privacy, that can undermine public trust and discourage uptake.
The public health benefit of these digital tools in outbreak responses needs to be further explored and better understood, especially for unintended consequences. In addition to the lack of evidence for real-world effectiveness, serious concerns remain that providing access to private information about health, behavior, and location can violate a user’s privacy—especially if these do not follow the critical principle of confidentiality and are repurposed for illegitimate surveillance purposes—as well as autonomy, if such technology is mandated (Gasser et al., 2020). Without deliberate investment and incentives to develop appropriate privacy preserving technologies for surveillance and contact tracing, this field will not advance as needed due to fear and ethical questions. Mistrust in these technologies would be a barrier to effective implementation and use. If digital technology is thus used at national or global scale during epidemics or pandemics, developing and instituting best practices and standards for responsible data collection and processing will be critical for engendering public trust (Ienca and Vayena, 2020). Moreover, many settings lack the capacity for local-, national-, and international-level data transmission and sharing through electronic platforms (Gao et al., 2020; Holmgren et al., 2020). Some empirical demonstration is beginning to support the potential real-world util-
ity of digital contact tracing during an infectious disease outbreak scenario, however. A population-based study looked at the impact of a digital contact tracing app implemented in the Canary Islands, Spain, during the summer of 2020 (Rodríguez et al., 2021). The app detected around six close contacts per simulated infection—most of whom were strangers—and the technology had relatively high adherence and compliance. Alongside these promising advances was a controversy regarding the United Kingdom’s National Health Service application, which, until mid-August 2021, could advise large groups and clusters of people to self-isolate. The use of this geolocator app placed worker shortages and continued COVID-19 related closures in conflict with the government’s wide-scale reopening plans (BBC, 2021).
SURVEILLANCE INNOVATIONS
The COVID-19 pandemic has underscored the need to broaden core capacities for surveillance by leveraging technological advances, including crowdsourced data streams, wastewater surveillance, metadata and line lists that link across data streams, and other innovative approaches. To strengthen preparedness and response to future epidemic and pandemic threats, these approaches—if determined to be effective and ethical—should be consolidated and routinized into central systems to complement traditional surveillance. Aligning international strategies for regulating, evaluating, and using digitally enabled public health is a key step toward realizing the full potential of public health in the future (Budd et al., 2020).
Wastewater Surveillance
The discovery that SARS-CoV-2 was present in infected patients’ feces and wastewater (Polo et al., 2020) has given rise to innovations in wastewater-based epidemiological surveillance that employ near-source tracking of sewage drains for specific buildings to detect individual cases or small clusters (Hassard et al., 2021). Wastewater surveillance for infectious diseases holds great potential value for population-wide monitoring and enabling detection of early signals of transmission dynamics, particularly when testing capacity is limited or the time to reporting of diagnostic test results is lengthy or delayed (Peccia et al., 2020). A study in Seattle, Washington, compared seven methods for concentrating and recovering SARS-CoV-2 from municipal wastewater and sludge (Philo et al., 2021). Skimmed milk flocculation without Vertrel extraction yielded the most consistent virus detection results and low variability, although the same may not hold in other contexts. Concentration and detection methods need to be appropriately validated for the setting’s specific water matrix to evaluate its performance.
Crowdsourcing Surveillance
Crowdsourcing surveillance by compiling lists of suspected, probable, and confirmed cases could enable quick preliminary assessments of epidemic growth, the potential for spread, appropriate periods of quarantine and isolation, and the efficiency of detection based on the current and evolving evidence base. A crowdsourcing approach was implemented in China in January 2020, when Kaiyuan Sun and colleagues compiled individual-level data from patients with COVID-19—which they mined from a Chinese social media network used by health care professionals—with province-level data about daily case counts (Leung and Leung, 2020). The information was synthesized into a crowdsourced line list that was well aligned with the official epidemiological reports released by the national government. In the future, such crowdsourcing strategies could help mitigate the spread of epidemics, dispel misinformation, and counteract the detrimental impacts of geopolitical tensions and nationalistic populations on science-based epidemic control efforts (Leung and Leung, 2020).
Rapid Epidemic Intelligence
Rapid epidemic intelligence draws on open-source data (e.g., news reports, social media) to supplement traditional surveillance methods and enable early detection of epidemic signals, thus supporting early investigation and accelerating the development of diagnostics. Algorithms for clinical syndromes or diseases, machine learning, and artificial intelligence can be used to establish a baseline threshold for detecting abnormal signals. For instance, an open-source epidemic observatory, EpiWatch, was able to detect early signals of pneumonia or severe acute respiratory illness as a proxy for COVID-19 at the outset of the pandemic in China and Indonesia (Kpozehouen et al., 2020; Thamtono et al., 2021). Similar sources of rapid epidemic intelligence include ProMED, Healthmap in the United States, the Global Public Health Intelligence Network (GPHIN) in Canada (Carter et al., 2020), and Epiwabak in Malaysia.
Nowcasting Surveillance
“Nowcasting” is an innovative framework for assessing the current state of an ongoing outbreak or epidemic by leveraging advances in computational and laboratory sciences to elucidate the event’s pathogenic, epidemiologic, clinical, and sociobehavioral characteristics; this approach can enhance situational awareness and inform decisions about response efforts (Wu et al., 2021).
Other Surveillance Innovations
Other surveillance-related innovations during COVID-19 have demonstrated success and feasibility. These include innovative partnerships, such as the COVID-19 Healthcare Coalition,2 data aggregation networks (Budd et al., 2020), blockchain technologies (Idrees et al., 2021; Mashamba-Thompson and Crayton, 2020), artificial intelligence surveillance tools (Allam et al., 2020), and pooled testing (FDA, 2020). Web-based dashboards can serve as dynamic tools for communicating data, informing decision making, and encouraging behavior change (Ivanković et al., 2021). Harvard has developed a smartphone app that detects loss of taste and smell, a strong indicator of COVID-19 (Hassard et al., 2021). WHO has explored using dogs to screen for COVID-19 (WHO, 2021a).
CONCLUSIONS AND RECOMMENDATIONS
Detection of Potential Threats
Conclusion: COVID-19 has further emphasized the need to use the One Health approach to better target surveillance, including by building on currently existing platforms for influenza surveillance in wild birds, poultry, and livestock. This includes programs for detection of new zoonotic strains with pandemic potential and large antigenic drifts and shifts and research to better understand the pandemic potential of new strains.
Conclusion: One Health programs need to identify new viral strains, assess the risk they pose to people, and analyze where cases are likely to be found and outbreaks are likely to begin. Interdisciplinary collaboration among U.S. agencies, academic institutions, national governments, and multilateral partners has been successful in performing this surveillance in several countries with a One Health approach.
Recommendation 2-1: The World Health Organization, the World Bank, and regional public health organizations should work collaboratively with countries (particularly low- and middle-income countries and those with extensive animal–human interfaces) to build sustainable capacity for routine surveillance in animals (wildlife, livestock, and domestic) and to develop and support interagency One Health platforms.
___________________
2 For more on the COVID-19 Healthcare Coalition, see https://dsd.c19hcc.org (accessed August 20, 2021).
Quantifying the Spread of a Pandemic
Conclusion: Data informing public health surveillance, including for influenza and COVID-19, are vulnerable to ascertainment biases and therefore may not reflect the true underlying epidemiology; these biases happen particularly as a novel strain is first emerging. When the means used to collect data cannot be changed to avoid these problems, they can be taken into consideration during the analysis and interpretation of data being used to inform policy decisions. If not corrected, these biases can misinform the public about a disease’s impact and the likely effects of public health interventions in general and in particular subgroups.
Conclusion: Within countries, the sharing of data collected from community-based surveillance is critical for identifying the likely impact of outbreaks. Inefficiencies in collecting and sharing all types and sources of data among countries and global health agencies hampered the flow of information during the COVID-19 pandemic. The rapid sharing of a wide range of data internationally, including syndromic, epidemiologic, clinical, pathogen specific (e.g., genomic), and other (such as open-source intelligence), can provide early warning of an outbreak of concern as well as a picture of how it may develop.
Recommendation 2-2: Countries should institute surveillance as the backbone of their health care systems, which should include submitting aggregated clinical data feeding into public health agencies. To ensure that policy makers have access to accurate, timely, and comprehensive risk assessments, national authorities—with the advice and assistance of regional and global public health agencies—should establish more robust surveillance systems, involving public hospitals and academic medical centers, manufacturers of diagnostics, and social network platforms. Epidemiologists should be alert to potential ascertainment biases regarding sampling frames and other methodological pitfalls, account for such biases during analysis and interpretation of the data, notify authorities to take these biases into account, and seek support for improving surveillance methods to better achieve representativeness and sufficient geographical coverage.
Tracing the Arrival and Community Transmission of a Virus
Conclusion: COVID-19 showed that countries and intergovernmental bodies need to bolster their surveillance capacities, especially the ability to look for the unexpected and unobserved and to sustain surveillance during disease surges. These systems can be strengthened
by being repeatedly challenged to assess their ability to detect novel threats. Gaps identified can then be followed through and retested iteratively before an actual incident. Current surveillance approaches and tools are designed and more suitable for monitoring of known pandemics or the ongoing surveillance for seasonal influenza than for the early detection of a pandemic-capable pathogen before widespread transmission.
Conclusion: COVID-19 showed that the set of core capacities should be broadened to take advantage of technological developments, including but not limited to, digital mobility data, sewage surveillance, and monitoring of open-access electronic data streams (digital surveillance), as well as to maintain a stockpile of basic supplies (such as nasal swabs) that will be needed to conduct tests).
Full reporting of surveillance data, both to higher authorities within a country and to international agencies, is sometimes impeded by negative political or economic repercussions. For example, disciplining local officials for reporting novel pathogens disincentivizes health surveillance. The first step in eliminating such barriers is to recognize their existence; such recognition can come from the parties involved or from observers. Unless such barriers are removed, reporting structures cannot provide complete, accurate, and timely information about possible disease outbreaks.
Harmonization of information from multiple data sources is essential for quickly identifying the origins and spread of novel agents and strains and for providing useful information for decision makers and the public. Harmonization rests on the development and use of instruments to standardize the data. When diverse data come from many sources and reflect clinical and public health differences at the local level, particularly in the early stages of a pandemic, organizations that collect the data may be able to develop means of standardizing the data after they have been submitted.
Recommendation 2-3: National public health agencies should both strengthen the capabilities of local and provincial authorities to accurately, rapidly, and transparently report data about novel agents and strains and improve their own reporting of data to such regional organizations and global bodies as the World Health Organization and the One Health Tripartite. The global bodies should develop methods to harmonize data from multiple sources, to enable prompt dissemination of useful, comprehensive data, especially to the national and regional organizations that have contributed to the data pool. Organizations to
which data are submitted at all levels should work toward removing barriers and disincentives to making full and accurate reports.
Recommendation 2-4: The World Health Organization and regional disease control agencies (e.g., European Centre for Disease Prevention and Control, Africa Centres for Disease Control and Prevention) should work with countries, and national governments should work with subnational entities (counties, states, provinces), to harmonize, coordinate, and optimize surveillance activities, data collection, and sharing.
REFERENCES
Abbey, E. J., B. A. Khalifa, M. O. Oduwole, S. K. Ayeh, R. D. Nudotor, E. L. Salia, O. Lasisi, S. Bennett, H. E. Yusuf, and A. L. Agwu. 2020. The Global Health Security Index is not predictive of coronavirus pandemic responses among Organizations for Economic Cooperation and Development countries. PLOS ONE 15(10):e0239398.
Aiello, A. E., A. Renson, and P. N. Zivich. 2020. Social media- and internet-based disease surveillance for public health. Annual Review of Public Health 41:101–118.
Aitken, T., K. L. Chin, D. Liew, and R. Ofori-Asenso. 2020. Rethinking pandemic preparation: Global health security index (GHSI) is predictive of COVID-19 burden, but in the opposite direction. The Journal of Infection 81(2):318–356.
Allam, Z., G. Dey, and D. S. Jones. 2020. Artificial intelligence (AI) provided early detection of the coronavirus (COVID-19) in China and will influence future urban health policy internationally. AI 1(2):156–165.
Alwan, N. A. 2020. Surveillance is underestimating the burden of the COVID-19 pandemic. The Lancet 396(10252):e24.
Angelopoulos, A. N., R. Pathak, R. Varma, and M. I. Jordan. 2020. On identifying and mitigating bias in the estimation of the COVID-19 case fatality rate. https://assets.pubpub.org/zeiuy9aq/71594236940800.pdf (accessed June 25, 2021).
Anglemyer, A., T. H. M. Moore, L. Parker, T. Chambers, A. Grady, K. Chiu, M. Parry, M. Wilczynska, E. Flemyng, and L. Bero. 2020. Digital contact tracing technologies in epidemics: A rapid review. Cochrane Database of Systematic Reviews (8).
Baum, S. E., C. Machalaba, P. Daszak, R. H. Salerno, and W. B. Karesh. 2017. Evaluating One Health: Are we demonstrating effectiveness? One Health (3):5–10.
BBC. 2021. Pingdemic: “We got close to complete shutdown.” BBC. 2021.
Bendavid, E., B. Mulaney, N. Sood, S. Shah, R. Bromley-Dulfano, C. Lai, Z. Weissberg, R. Saavedra-Walker, J. Tedrow, A. Bogan, T. Kupiec, D. Eichner, R. Gupta, J. P. A. Ioannidis, and J. Bhattacharya. 2021. COVID-19 antibody seroprevalence in Santa Clara County, California. International Journal of Epidemiology 50(2):410–419
Bogich, T. L., R. Chunara, D. Scales, E. Chan, L. C. Pinheiro, A. A. Chmura, D. Carroll, P. Daszak, and J. S. Brownstein. 2012. Preventing pandemics via international development: A systems approach. PLOS Medicine 9(12):e1001354.
Bradshaw, W. J., E. C. Alley, J. H. Huggins, A. L. Lloyd, and K. M. Esvelt. 2021. Bidirectional contact tracing could dramatically improve COVID-19 control. Nature Communications 12(1):232.
Brammer, L., L. Blanton, S. Epperson, D. Mustaquim, A. Bishop, K. Kniss, R. Dhara, M. Nowell, L. Kamimoto, and L. Finelli. 2011. Surveillance for influenza during the 2009 influenza A (H1N1) pandemic—United States, April 2009–March 2010. Clinical Infectious Diseases 52(Suppl 1):S27–S35.
Budd, J., B. S. Miller, E. M. Manning, V. Lampos, M. Zhuang, M. Edelstein, G. Rees, V. C. Emery, M. M. Stevens, N. Keegan, M. J. Short, D. Pillay, E. Manley, I. J. Cox, D. Heymann, A. M. Johnson, and R. A. McKendry. 2020. Digital technologies in the public-health response to COVID-19. Nature Medicine 26(8):1183–1192.
Cancryn, A. 2020. How overly optimistic modeling distorted Trump team’s coronavirus response. https://www.politico.com/news/2020/04/24/trump-coronavirus-model-207582 (accessed June 25, 2021).
Carroll, D., S. Morzaria, S. Briand, C. K. Johnson, D. Morens, K. Sumption, O. Tomori, and S. Wacharphaueasadee. 2021. Preventing the next pandemic: The power of a global viral surveillance network. BMJ 372:n485.
Carter, D., M. Stojanovic, P. Hachey, K. Fournier, S. Rodier, Y. Wang, and B. De Bruijn. 2020. Global public health surveillance using media reports: Redesigning GPHIN. arXiv (preprint):2004.04596.
CDC (U.S. Centers for Disease Control and Prevention). 2020. U.S. influenza surveillance system: Purpose and methods. https://www.cdc.gov/flu/weekly/overview.htm (accessed June 25, 2021).
CDC. 2021a. Genomic surveillance for SARS-COV-2 variants. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance.html (accessed April 18, 2021).
CDC. 2021b. Influenza virus genome sequencing and genetic characterization. https://www.cdc.gov/flu/about/professionals/genetic-characterization.htm (accessed June 21, 2021).
Chan, J. F., S. Yuan, K. H. Kok, K. K. To, H. Chu, J. Yang, F. Xing, J. Jiu, C. C. Yip, R. W. Poon, H. Tsoi, S. K. Lo, K. Chan, V. K. Poon, W. Chan, J. Daniel, J. Cai, V. C. Cheng, H. Chen, C. K. Hui, and K. Yuen. 2020. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. The Lancet 395(10223):514–523.
Chow, C. C., J. C. Chang, R. C. Gerkin, and S. Vattikuti. 2020. Global prediction of unreported SARS-CoV-2 infection from observed COVID-19 cases. medRxiv 2020.04.29.20083485.
Chu, A., R. Savage, D. Willison, N. S. Crowcroft, L. C. Rosella, D. Sider, J. Garay, I. Gemmill, A.-L. Winter, R. F. Davies, and I. Johnson. 2012. The use of syndromic surveillance for decision-making during the H1N1 pandemic: A qualitative study. BMC Public Health 12(1):929.
Chu, H. Y., J. A. Englund, L. M. Starita, M. Famulare, E. Brandstetter, D. A. Nickerson, M. J. Rieder, A. Adler, K. Lacombe, A. E. Kim, C. Graham, J. Logue, C. R. Wolf, J. Heimonen, D. J. McCulloch, P. D. Han, T. R. Sibley, J. Lee, M. Ilcisin, K. Fay, R. Burstein, B. Martin, C. M. Lockwood, M. Thompson, B. Lutz, M. Jackson, J. P. Hughes, M. Boeckh, J. Shendure, and T. Bedford. 2020. Early detection of COVID-19 through a citywide pandemic surveillance platform. New England Journal of Medicine 383(2):185–187.
Cosgriff, C. V., D. K. Ebner, and L. A. Celi. 2020. Data sharing in the era of COVID-19. The Lancet Digital Health 2(5):e224.
de Lusignan, S., N. Jones, J. Dorward, R. Byford, H. Liyanage, J. Briggs, F. Ferreira, O. Akinyemi, G. Amirthalingam, C. Bates, J. Lopez Bernal, G. Dabrera, A. Eavis, A. J. Elliot, M. Feher, E. Krajenbrink, U. Hoang, G. Howsam, J. Leach, C. Okusi, B. Nicholson, P. Nieri, J. Sherlock, G. Smith, M. Thomas, N. Thomas, M. Tripathy, W. Victor, J. Williams, I. Wood, M. Zambon, J. Parry, S. O’Hanlon, M. Joy, C. Butler, M. Marshall, and F. D. R. Hobbs. 2020. The Oxford Royal College of General Practitioners clinical informatics digital hub: Protocol to develop extended COVID-19 surveillance and trial platforms. JMIR Public Health Surveillance 6(3):e19773.
ECDC (European Centre for Disease Prevention and Control). 2020. Options for the use of rapid antigen tests for COVID-19 in the EU/EEA and the UK. https://www.ecdc.europa.eu/sites/default/files/documents/Options-use-of-rapid-antigen-tests-for-COVID-19.pdf (accessed June 25, 2021).
ECDC. 2021. Sentinel surveillance. https://www.ecdc.europa.eu/en/seasonal-influenza/surveillance-and-disease-data/facts-sentinel-surveillance (accessed June 25, 2021).
Elliot, A. J., S. E. Harcourt, H. E. Hughes, P. Loveridge, R. A. Morbey, S. Smith, A. Soriano, A. Bains, G. E. Smith, O. Edeghere, and I. Oliver. 2020. The COVID-19 pandemic: A new challenge for syndromic surveillance. Epidemiology and Infection 148:e122.
Elton, L., N. Haider, R. Kock, M. J. Thomason, J. Tembo, L. B. Arruda, F. Ntoumi, A. Zumla, and T. D. McHugh. 2021. Zoonotic disease preparedness in Sub-Saharan African countries. One Health Outlook 3(1):5.
Ending Pandemics. 2021. PODD (Participatory One Health Digital Disease Detection). https://endingpandemics.org/projects/participatory-one-health-digital-disease-detection-podd (accessed June 17, 2021).
FDA (U.S. Food and Drug Administration). 2020. Coronavirus (COVID-19) update: Facilitating diagnostic test availability for asymptomatic testing and sample pooling. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-facilitating-diagnostic-test-availability-asymptomatic-testing-and (accessed April 18, 2021).
Fiore, V. G., N. DeFelice, B. S. Glicksberg, O. Perl, A. Shuster, K. Kulkarni, M. O’Brien, M. A. Pisauro, D. Chung, and X. Gu. 2021. Containment of COVID-19: Simulating the impact of different policies and testing capacities for contact tracing, testing, and isolation. PLOS ONE 16(3):e0247614.
Fonseca, P. 2021. Brazil COVID-19 testing shrinks as cases and deaths accelerate. https://www.reuters.com/article/us-health-coronavirus-brazil-testing-idUSKBN2B423E (accessed June 17, 2021).
Frimpong, J. A., M. M. Park, M. P. Amo-Addae, P. A. Adewuyi, and T. K. Nagbe. 2017. Detecting, reporting, and analysis of priority diseases for routine public health surveillance in Liberia. The Pan African Medical Journal 27(Suppl 1):9.
Fukushima, M., C. Austin, N. Sato, T. Maruyama, E. Navarro, M. Rocca, J. Demotes, M. Haendel, S. L. Volchenboum, M. Cowperthwaite, J. C. Silverstein, C. Webb, I. Sim, M. Chase, J. Speakman, E. Augustine, D. E. Ford, and R. Kush. 2018. The global academic research organization network: Data sharing to cure diseases and enable learning health systems. Learning Health Systems 3:e10073. https://doi.org/10.1002/lrh2.10073.
Gao, F., L. Tao, Y. Huang, and Z. Shu. 2020. Management and data sharing of COVID-19 pandemic information. Biopreservation and Biobanking 18(6):570–580.
Garg, S., L. Kim, M. Whitaker, A. O’Halloran, C. Cummings, R. Holstein, M. Prill, S. J. Chai, P. D. Kirley, N. B. Alden, B. Kawasaki, K. Yousey-Hindes, L. Niccolai, E. J. Anderson, K. P. Openo, A. Weigel, M. L. Monroe, P. Ryan, J. Henderson, S. Kim, K. Como-Sabetti, R. Lynfield, D. Sosin, S. Torres, A. Muse, N. M. Bennett, L. Billing, M. Sutton, N. West, W. Schaffner, H. K. Talbot, C. Aquino, A. George, A. Budd, L. Brammer, G. Langley, A. J. Hall, and A. Fry. 2020. Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019-COVID-net, 14 states, March 1–30, 2020. Morbidity and Mortality Weekly Report 69(15):458–464.
Gasser, U., M. Ienca, J. Scheibner, J. Sleigh, and E. Vayena. 2020. Digital tools against COVID-19: Taxonomy, ethical challenges, and navigation aid. The Lancet Digital Health 2(8):e425–e434.
Ge, X. Y., J. L. Li, X. L. Yang, A. A. Chmura, G. Zhu, J. H. Epstein, J. K., Mazet, B. Hu, W. Zhang, C. Peng, Y. J. Zhang, C. M. Luo, B. Tan, N. Wang, Y. Zhu, B. Crameri, S. Y. Zhang, L. F. Wang, P. Daszak, and Z. L. Shi. 2013. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503(7477):535–538.
Goodman, J. D., and M. Rothfeld. 2020. 1 in 5 New Yorkers may have had COVID-19, antibody tests suggest. https://www.nytimes.com/2020/04/23/nyregion/coronavirus-antibodies-test-ny.html (accessed June 25, 2021).
Grange, Z. L., T. Goldstein, C. K. Johnson, S. Anthony, K. Gilardi, P. Daszak, K. J. Olival, T. O’Rourke, S. Murray, S. H. Olson, E. Togami, G. Vidal, and J. A. K. Mazet. 2021. Ranking the risk of animal-to-human spillover for newly discovered viruses. Proceedings of the National Academy of Sciences of the United States of America 118(15):e2002324118.
Haider, N., A. Yavlinsky, Y.-M. Chang, M. N. Hasan, C. Benfield, A. Y. Osman, M. J. Uddin, O. Dar, F. Ntoumi, A. Zumla, and R. Kock. 2020. The Global Health Security Index and Joint External Evaluation score for health preparedness are not correlated with countries’ COVID-19 detection response time and mortality outcome. Epidemiology and Infection 148:e210.
Hassard, F., L. Lundy, A. C. Singer, J. Grimsley, and M. Di Cesare. 2021. Innovation in wastewater near-source tracking for rapid identification of COVID-19 in schools. The Lancet Microbe 2(1):e4–e5.
Hellewell, J., S. Abbott, A. Gimma, N. I. Bosse, C. I. Jarvis, T. W. Russell, J. D. Munday, A. J. Kucharski, W. J. Edmunds, F. Sun, S. Flasche, B. J. Quilty, N. Davies, Y. Liu, S. Clifford, P. Klepac, M. Jit, C. Diamond, H. Gibbs, K. van Zandvoort, S. Funk, and R. M. Eggo. 2020. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. The Lancet Global Health 8(4):e488–e496.
Holmdahl, I., and C. Buckee. 2020. Wrong but useful—what COVID-19 epidemiologic models can and cannot tell us. New England Journal of Medicine 383(4):303–305.
Holmgren, A. J., N. C. Apathy, and J. Adler-Milstein. 2020. Barriers to hospital electronic public health reporting and implications for the COVID-19 pandemic. Journal of the American Medical Informatics Association 27(8):1306–1309.
Huong, N. Q., N. T. T. Nga, N. V. Long, B. D. Luu, A. Latinne, M. Pruvot, N. T. Phuong, L. T. V. Quang, V. V. Hung, N. T. Lan, N. T. Hoa, P. Q. Minh, N. T. Diep, N. Tung, V. D. Ky, S. I. Roberton, H. B. Thuy, N. V. Long, M. Gilbert, L. Wicker, J. A. K. Mazet, C. K. Johnson, T. Goldstein, A. Tremeau-Bravard, V. Ontiveros, D. O. Joly, C. Walzer, A. E. Fine, and S. H. Olson. 2020. Coronavirus testing indicates transmission risk increases along wildlife supply chains for human consumption in Vietnam, 2013–2014. PLOS ONE 15(8):e0237129.
Idrees, S. M., M. Nowostawski, and R. Jameel. 2021. Blockchain-based digital contact tracing apps for COVID-19 pandemic management: Issues, challenges, solutions, and future directions. JMIR Medical Informatics 9(2):e25245.
IEDCR (Institute of Epidemiology, Disease Control, and Research). 2012. Strategic framework for One Health approach to infectious diseases in Bangladesh. http://www.iedcr.org/pdf/files/One%20Health/Strategic_framework_for_One_Health_Bangladesh-26%20Jan.pdf (accessed August 30, 2021).
Ienca, M., and E. Vayena. 2020. On the responsible use of digital data to tackle the COVID-19 pandemic. Nature Medicine 26(4):463–464.
IHME (Institute for Health Metrics and Evaluation). 2020. Our COVID-19 forecasting model, otherwise known as “The Chris Murray Model.” http://www.healthdata.org/acting-data/our-covid-19-forecasting-model-otherwise-known-chris-murray-model (accessed June 25, 2021).
Inzaule, S. C., S. K. Tessema, Y. Kebede, A. E. Ogwell Ouma, and J. N. Nkengasong. 2021. Genomic-informed pathogen surveillance in Africa: Opportunities and challenges. The Lancet Infectious Diseases 21(9):E281–E289.
Ioannidis, J. P. 2021. Infection fatality rate of COVID-19 inferred from seroprevalence data. Bulletin of the World Health Organization 99(1):19.
IOM (Institute of Medicine). 2015. Emerging viral diseases: The One Health connection: Workshop summary. Washington, DC: The National Academies Press.
IOM and NRC (National Research Council). 2009. Sustaining global surveillance and response to emerging zoonotic diseases. Washington, DC: The National Academies Press.
Ivankovi ć, D., E. Barbazza, V. Bos, Ó. B. Fernandes, K. J. Gilmore, T. Jansen, P. Kara, N. Larrain, S. Lu, B. Meza-Torres, J. Mulyanto, M. Poldrugovac, A. Rotar, S. Wang, C. Willmington, Y. Yang, Z. Yelgezekova, S. Allin, N. Klazinga, and D. Kringos. 2021. Features constituting actionable COVID-19 dashboards: Descriptive assessment and expert appraisal of 158 public Web-based COVID-19 dashboards. Journal of Medical Internet Research 23(2).
Jewell, N. P., J. A. Lewnard, and B. L. Jewell. 2020. Caution warranted: Using the Institute for Health Metrics and Evaluation model for predicting the course of the COVID-19 pandemic. Annals of Internal Medicine 173(3):226–227.
JHU (Johns Hopkins University). 2019. Global health security index. https://www.ghsindex.org/wp-content/uploads/2019/10/2019-Global-Health-Security-Index.pdf (accessed June 17, 2021).
Kelly, T. R., C. Machalaba, W. B. Karesh, P. Z. Crook, K. Gilardi, J. Nziza, M. M. Uhart, E. A. Robles, K. Saylors, D. O. Joly, C. Monagin, P. M. Mangombo, P. M. Kingebeni, R. Kazwala, D. Wolking, W. Smith, PREDICT Consortium, and J. A. K. Mazet. 2020. Implementing One Health approaches to confront emerging and re-emerging zoonotic disease threats: Lessons from PREDICT. One Health Outlook (2):1.
Kobayashi, T., S.-M. Jung, N. M. Linton, R. Kinoshita, K. Hayashi, T. Miyama, A. Anzai, Y. Yang, B. Yuan, A. R. Akhmetzhanov, A. Suzuki, and H. Nishiura. 2020. Communicating the risk of death from novel coronavirus disease (COVID-19). Journal of Clinical Medicine 9(2):580.
Koonin, L. M., B. Hoots, C. A. Tsang, Z. Leroy, K. Farris, T. Jolly, P. Antall, B. McCabe, C. B. R. Zelis, I. Tong, and A. M. Harris. 2020. Trends in the use of telehealth during the emergence of the COVID-19 pandemic—United States, January–March 2020. Morbidity and Mortality Weekly Report 69(43):1595–1599.
Kpozehouen, E. B., X. Chen, M. Zhu, and C. R. Macintyre. 2020. Using open-source intelligence to detect early signals of COVID-19 in China: Descriptive study. JMIR Public Health and Surveillance 6(3):e18939.
Lancet Digital Health. 2020. Contact tracing: Digital health on the frontline. The Lancet Digital Health 2(11):e561.
Latinne, A., B. Hu, K. J. Olival, G. Zhu, L. Zhang, H. Li, A. A. Chmura, H. E. Field, C. Zambrana-Torrelio, J. H. Epstein, B. Li, W. Zhang, L. F. Wang, Z. L. Shi, and P. Daszak. 2020. Origin and cross-species transmission of bat coronaviruses in China. Nature Communications 11(1):4235.
Leung, G. M., and K. Leung. 2020. Crowdsourcing data to mitigate epidemics. The Lancet Digital Health 2(4):e156–e157.
Lipsitch, M., F. G. Hayden, B. J. Cowling, and G. M. Leung. 2009. How to maintain surveillance for novel influenza A H1N1 when there are too many cases to count. The Lancet 374(9696):1209–1211.
Liverani, M., S. Teng, M. S. Le, and R. Coker. 2018. Sharing public health data and information across borders: Lessons from Southeast Asia. Globalization and Health 14(1):94.
Loh, T. 2020. Germany loses control of contact tracing for COVID-19. https://www.bloomberg.com/news/articles/2020-10-28/even-germans-have-lost-control-of-tracing-as-infections-surge (accessed June 17, 2021).
LoTempio, J., R. Y. D’Andre Spencer, A. D. Vilain, E. Vilain, and E. Délot. 2020. We can do better: Lessons learned on data sharing in COVID-19 pandemic can inform future outbreak preparedness and response. Science & Diplomacy 9(2).
Mashamba-Thompson, T. P., and E. D. Crayton. 2020. Blockchain and artificial intelligence technology for novel coronavirus disease 2019 self-testing. Diagnostics 10(4):198.
Maxmen, A. 2021. Why U.S. coronavirus tracking can’t keep up with concerning variants. https://www.nature.com/articles/d41586-021-00908-0 (accessed August 28, 2021).
McKenzie, J. S., R. Dahal, M. Kakkar, N. Debnath, M. Rahman, S. Dorjee, K. Naeem, T. Wijayathilaka, B. K. Sharma, N. Maidanwal, A. Halimi, E. Kim, P. Chatterjee, and B. Devleesschauwer. 2016. One Health research and training and government support for One Health in South Asia. Infection Ecology & Epidemiology 6:1.
McLaughlin, H. P., B. C. Hiatt, D. Russell, C. M. Carlson, J. R. Jacobs, A. C. Perez-Osorio, M. L. Holshue, S. W. Choi, and R. K. Gautom. 2021. COVID-19 response efforts of Washington state public health laboratory: Lessons learned. American Journal of Public Health e1–e9.
Menachery, V. D., B. L. Yount, Jr., K. Debbink, S. Agnihothram, L. E. Gralinski, J. A. Plante, R. L. Graham, T. Scobey, X. Y. Ge, E. F. Donaldson, S. H. Randell, A. Lanzavecchia, W. A. Marasco, Z. L. Shi, and R. S. Baric. 2015. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nature Medicine 21(12):1508–1513.
Meyer, R., and A. C. Madrigal. 2021. We tracked America’s COVID-19 numbers for a year. The Atlantic. https://www.theatlantic.com/science/archive/2021/03/americas-coronavirus-catastrophe-began-with-data/618287 (accessed June 17, 2021).
Meyers, L., G. C. Ginocchio, A. N. Faucett, F. S. Nolte, P. H. Gesteland, A. Leber, D. Janowiak, V. Donovan, J. Dien Bard, S. Spitzer, K. A. Stellrecht, H. Salimnia, R. Selvarangan, S. Juretschko, J. A. Daly, J. C. Wallentine, K. Lindsey, F. Moore, S. L. Reed, M. Aguero-Rosenfeld, P. D. Fey, G. A. Storch, S. J. Melnick, C. C. Robinson, J. F. Meredith, C. V. Cook, R. K. Nelson, J. D. Jones, S. V. Scarpino, B. M. Althouse, K. M. Ririe, B. A. Malin, and M. A. Poritz. 2018. Automated real-time collection of pathogen-specific diagnostic data: Syndromic infectious disease epidemiology. JMIR Public Health and Surveillance 4(3):e59.
Mina, M. J., and K. G. Andersen. 2021. COVID-19 testing: One size does not fit all. Science 371(6525):126–127.
Morgan, O., X. Aguilera, A. Ammon, J. Amuasi, I. S. Fall, T. Frieden, D. Heymann, C. Ihekweazu, E. Jeong, G. M. Leung, B. Mahon, J. Nkengasong, F. N. Qamor, A. Schuchat, L. H. Wieler, and S. F. Dowell. 2021. Disease surveillance for the COVID-19 era: Time for bold changes. The Lancet 397(10292):2317–2319.
Munyua, P. M., M. K. Njenga, E. M. Osoro, C. O. Onyango, A. O. Bitek, A. Mwatondo, M. K. Muturi, N. Musee, G. Bigogo, E. Otiang, F. Ade, S. A. Lowther, R. F. Breiman, J. Neatherlin, J. Montgomery, and M.-A. Widdowson. 2019. Successes and challenges of the One Health approach in Kenya over the last decade. BMC Public Health 19(3):465.
Nachega, J. B., R. Atteh, C. Ihekweazu, N. A. Sam-Agudu, P. Adejumo, S. Nsanzimana, E. Rwagasore, J. Condo, M. Paleker, H. Mahomed, F. Suleman, A. R. Ario, E. Kiguli-Malwadde, F. G. Omaswa, N. K. Sewankambo, C. Viboud, M. J. A. Reid, A. Zumla, and P. H. Kilmarx. 2021. Contact tracing and the COVID-19 response in Africa: Best practices, key challenges, and lessons learned from Nigeria, Rwanda, South Africa, and Uganda. American Journal of Tropical Medicine and Hygiene 104(4):1179–1187.
NBC New York. 2020. “No Time to Be Lax”: Cuomo extends New York shutdown, NJ deaths top 1,000. https://www.nbcnewyork.com/news/local/our-9-11-moment-new-york-braces-as-nations-top-doctor-warns-saddest-week-ahead/2361336 (accessed June 25, 2021).
Ng, Y., Z. Li, Y. X. Chua, W. L. Chaw, Z. Zhao, B. Er, R. Pung, C. J. Chiew, D. C. Lye, and D. Heng. 2020. Evaluation of the effectiveness of surveillance and containment measures for the first 100 patients with COVID-19 in Singapore—January 2–February 29, 2020. Morbidity and Mortality Weekly Report 69(11):307–311.
NYC (New York City) Planning. 2021. Population–New York City population. https://www1.nyc.gov/site/planning/planning-level/nyc-population/population-facts.page (accessed September 21, 2021).
Okello, A. L., K. Bardosh, J. Smith, and S. C. Welburn. 2014. One Health: Past successes and future challenges in three African contexts. PLOS Neglected Tropical Diseases 8(5):e2884.
Oleribe, O. O., M. M. Crossey, and S. D. Taylor-Robinson. 2015. Nigerian response to the 2014 Ebola viral disease outbreak: Lessons and cautions. The Pan African Medical Journal 22(Suppl 1):13.
Pavelka, M., K. Van Zandvoort, S. Abbott, K. Sherratt, M. Majdan, CMMID COVID-19 working group; Inštitút Zdravotných Analýz, P. Jarčuška, M. Krajčí, and S. Flasche. 2021. The impact of population-wide rapid antigen testing on SARS-CoV-2 prevalence in Slovakia. Science 372(6542):635–641.
Peccia, J., A. Zulli, D. E. Brackney, N. D. Grubaugh, E. H. Kaplan, A. Casanovas-Massana, A. I. Ko, A. A. Malik, D. Wang, M. Wang, J. L. Warren, D. M. Weinberger, W. Arnold, and S. B. Omer. 2020. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nature Biotechnology 38(10):1164–1167.
Peiris, J. S. M., B. J. Cowling, J. T. Wu, L. Feng, Y. Guan, H. Yu, and G. M. Leung. 2016. Interventions to reduce zoonotic and pandemic risks from avian influenza in Asia. The Lancet Infectious Diseases 16(2):252–258.
Peixoto, V. R., C. Nunes, and A. Abrantes. 2020. Epidemic surveillance of COVID-19: Considering uncertainty and under-ascertainment. Portuguese Journal of Public Health 38(1):23–29.
Petrikova, I., J. Cole, and A. Farlow. 2020. COVID-19, wet markets, and planetary health. Lancet Planetary Health 4(6):e213–e214.
Philo, S. E., E. K. Keim, R. Swanstrom, A. Q. W. Ong, E. A. Burnor, A. L. Kossik, J. C. Harrison, B. A. Demeke, N. A. Zhou, N. K. Beck, J. H. Shirai, and J. S. Meschke. 2021. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Science of the Total Environment 760:144215.
Pisani, E., and C. AbouZahr. 2010. Sharing health data: good intentions are not enough. Bulletin of the World Health Organization 88(6):462–466. doi: 10.2471/BLT.09.074393. PMID: 20539861. PMCID: PMC2878150.
Pollán, M., B. Pérez-Gómez, R. Pastor-Barriuso, J. Oteo, M. A. Hernán, M. Pérez-Olmeda, J. L. Sanmartín, A. Fernández-García, I. Cruz, and N. F. de Larrea. 2020. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): A nationwide, population-based seroepidemiological study. The Lancet 396(10250):535–544.
Polo, D., M. Quintela-Baluja, A. Corbishley, D. L. Jones, A. C. Singer, D. W. Graham, and J. L. Romalde. 2020. Making waves: Wastewater-based epidemiology for COVID-19—Approaches and challenges for surveillance and prediction. Water Research 186:116404.
Reddin, K., H. Bang, and L. Miles. 2021. Evaluating simulations as preparation for health crises like COVID-19: Insights on incorporating simulation exercises for effective response. The International Journal of Disaster Risk Reduction 102245.
Reintjes, R. 2020. Lessons in contact tracing from Germany: Germany built on existing local infrastructure to get ahead of the COVID-19 pandemic. The BMJ 369.
Rodríguez, P., S. Graña, E. E. Alvarez-León, M. Battaglini, F. J. Darias, M. A. Hernán, R. López, P. Llaneza, M. C. Martín, O. Ramirez-Rubio, A. Romaní, B. Suárez-Rodríguez, J. Sánchez-Monedero, A. Arenas, L. Lacasa, and RadarCovidPilot Group. 2021. A population-based controlled experiment assessing the epidemiological impact of digital contact tracing. Nature Communications 12(1):587.
Salathé, M., C. L. Althaus, N. Anderegg, D. Antonioli, T. Ballauz, B. Edouard, S. Čapkun, D. Jackson, S.-I. Kim, J. R. Larus, N. Low, W. Lueks, D. Menges, C. Moullet, M. Payer, J. Riou, T. Stadler, C. Troncoso, E. Vayena, and V. V. Wyl. 2020. Early evidence of effectiveness of digital contact tracing for SARS-CoV-2 in Switzerland. medRxiv 2020.2009.2007.20189274.
SARInet (Severe Acute Respiratory Infections Network). 2021. Explore SARInet. https://www.sarinet.org/en/explore-sarinet (accessed June 25, 2021).
Seto, J., Y. Aoki, K. Komabayashi, Y. Ikeda, M. Sampei, N. Ogawa, Y. Uchiumi, S. Fujii, M. Chiba, E. Suzuki, T. Takahashi, K. Yamada, Y. Otani, Y. Ashino, K. Araki, T. Kato, H. Ishikawa, T. Ikeda, H. Abe, T. Ahiko, and K. Mizuta. 2021. Epidemiology of coronavirus disease 2019 in Yamagata prefecture, Japan, January–May 2020: The importance of retrospective contact tracing. Japanese Journal of Infectious Diseases.
Shaman, J., E. Goldstein, and M. Lipsitch. 2011. Absolute humidity and pandemic versus epidemic influenza. American Journal of Epidemiology 173(2):127–135.
Sharfstein, J. M., S. J. Becker, and M. M. Mello. 2020. Diagnostic testing for the novel coronavirus. JAMA 323(15):1437–1438.
Silverman, J. D., N. Hupert, and A. D. Washburne. 2020. Using influenza surveillance networks to estimate state-specific prevalence of SARS-CoV-2 in the United States. Science Translational Medicine 12(554):eabc1126.
Skrip, L. A., P. Selvaraj, B. Hagedorn, A. L. Ouédraogo, N. Noori, A. Orcutt, D. Mistry, J. Bedson, L. Hébert-Dufresne, S. V. Scarpino, and B. M. Althouse. 2021. Seeding COVID-19 across Sub-Saharan Africa: An analysis of reported importation events across 49 countries. American Journal of Tropical Medicine and Hygiene 104(5):1694–1702.
Thamtono, Y., A. Moa, and C. R. MacIntyre. 2021. Using open-source intelligence to identify early signals of COVID-19 in Indonesia. Western Pacific Surveillance and Response 12(1):6.
Thielen, P. M., S. Wohl, T. Mehoke, S. Ramakrishnan, M. Kirsche, O. Falade-Nwulia, N. S. Trovão, A. Ernlund, C. Howser, N. Sadowski, C. P. Morris, M. Hopkins, M. Schwartz, Y. Fan, V. Gniazdowski, J. Lessler, L. Sauer, M. C. Schatz, J. D. Evans, S. C. Ray, W. Timp, and H. H. Mostafa. 2021. Genomic diversity of SARS-CoV-2 during early introduction into the Baltimore-Washington metropolitan area. JCI Insight 6(6):e144350.
Tiwari, R., K. Dhama, K. Sharun, M. Iqbal Yatoo, Y. S. Malik, R. Singh, I. Michalak, R. Sah, D. K. Bonilla-Aldana, and A. J. Rodriguez-Morales. 2020. COVID-19: Animals, veterinary and zoonotic links. Veterinary Quarterly 40(1):169–182.
Tsang, T. K., P. Wu, Y. Lin, E. H. Y. Lau, G. M. Leung, and B. J. Cowling. 2020. Effect of changing case definitions for COVID-19 on the epidemic curve and transmission parameters in mainland China: A modelling study. Lancet Public Health 5(5):e289–e296.
Tsao, S.-F., H. Chen, T. Tisseverasinghe, Y. Yang, L. Li, and Z. A. Butt. 2021. What social media told us in the time of COVID-19: A scoping review. Lancet Digital Health 3(3):e175–e194.
United Nations Development Programme China. 2017. 2017 Report on the sustainable development of Chinese enterprises overseas. https://www.cn.undp.org/content/china/en/home/library/south-south-cooperation/2017-report-on-the-sustainable-development-of-chinese-enterprise.html (accessed August 30, 2021).
van den Wijngaard, C., L. van Asten, W. van Pelt, N. J. D. Nagelkerke, R. Verheij, A. J. de Neeling, A. Dekkers, M. A. B. van der Sande, H. van Vliet, and M. P. G. Koopmans. 2008. Validation of syndromic surveillance for respiratory pathogen activity. Emerging Infectious Diseases 14(6):917–925.
Verity, R., L. C. Okell, I. Dorigatti, P. Winskill, C. Whittaker, N. Imai, G. Cuomo-Dannenburg, H. Thompson, P. G. Walker, and H. Fu. 2020. Estimates of the severity of COVID-19 disease. medRxiv 20(6):669–677.
Vicentini, C., S. Bazzolo, D. Gamba, and C. M. Zotti. 2020. Analysis of the fatality rate in relation to testing capacity during the first 50 days of the COVID-19 epidemic in Italy. American Journal of Tropical Medicine and Hygiene 103(6):2382–2390.
Wang, N., S. Y. Li, X. L. Yang, H. M. Huang, Y. J. Zhang, H. Guo, C. M. Luo, M. Miller, G. J. Zhu, A. A. Chmura, E. Hagan, J. H. Zhou, Y. Z. Zhang, L. F. Wang, P. Daszak, and Z. L. Shi. 2018. Serological evidence of bat SARS-related coronavirus infection in humans, China. Virologica Sinica 33:104–107.
Wilmes, P., J. Zimmer, J. Schulz, F. Glod, L. Veiber, L. Mombaerts, B. Rodrigues, A. Aalto, J. Pastore, and C. J. Snoeck. 2021. SARS-CoV-2 transmission risk from asymptomatic carriers: Results from a mass screening programme in Luxembourg. The Lancet Regional Health-Europe 4:100056.
Winters, M., B. Oppenheim, J. Pick, and H. Nordenstedt. 2020. Creating misinformation: How a headline in the BMJ about COVID-19 spread virally. BMJ 369.
WHO (World Health Organization). 2021a. Consultation on the use of trained dogs for screening COVID-19 cases. https://cdn.who.int/media/docs/default-source/blue-print/who-consultation-screening-dogs--8th-march-20215d9ed791-3201-4e3c-93c1-a846910d53cc.pdf?sfvrsn=202ce0a_1&download=true (accessed April 18, 2021).
WHO. 2021b. WHO-convened global study of origins of SARS-CoV-2: China part. https://www.who.int/publications/i/item/who-convened-global-study-of-origins-of-sars-cov-2-china-part (accessed November 19, 2021).
WHO. 2021c. Global influenza surveillance and response system. https://www.who.int/initiatives/global-influenza-surveillance-and-response-system (accessed June 25, 2021).
Wu, J. T., K. Leung, M. Bushman, N. Kishore, R. Niehus, P. M. de Salazar, B. J. Cowling, M. Lipsitch, and G. M. Leung. 2020. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nature Medicine 26(4):506–510.
Wu, J. T., K. Leung, T. T. Y. Lam, M. Y. Ni, C. K. H. Wong, J. S. M. Peiris, and G. M. Leung. 2021. Nowcasting epidemics of novel pathogens: Lessons from COVID-19. Nature Medicine 27(3):388–395.
Xu, B., B. Gutierrez, S. Mekaru, K. Sewalk, L. Goodwin, A. Loskill, E. L. Cohn, Y. Hswen, S. C. Hill, M. M. Cobo, A. E. Zarebski, S. Li, C.-H. Wu, E. Hulland, J. D. Morgan, L. Wang, K. O’Brien, S. V. Scarpino, J. S. Brownstein, O. G. Pybus, D. M. Pigott, and M. U. G. Kraemer. 2020. Epidemiological data from the COVID-19 outbreak, real-time case information. Scientific Data 7(1):106.
Yalaman, A., G. Basbug, C. Elgin, and A. P. Galvani. 2021. Cross-country evidence on the association between contact tracing and COVID-19 case fatality rates. Scientific Reports 11(1):2145.




































