4
Strengthening Surveillance
Surveillance is an essential public health service. The information gained through the monitoring of a health threat, in this case antimicrobial resistance, informs public policy and communication; it also helps direct research attention (OPHSS and CDC, 2018). In this discussion, surveillance refers to the timely collection, analysis, and communication of data on resistance patterns (i.e., the extent, spread, evolution, and impact) for pathogens of public health importance (WHO, 2014). Improving surveillance is a key component of global and national action plans for combating antimicrobial resistance (Ranjalkar and Chandy, 2019; The White House, 2015; WHO, 2014).
Monitoring resistance is also a component of some disease surveillance programs. Resistance to the drugs that treat tuberculosis is an important part of national tuberculosis surveillance, for example. But unlike disease surveillance systems, surveillance for antimicrobial resistance requires monitoring a range of targets not just in human health, but in animals, crops, and the environment as well. The targets for monitoring include resistant pathogens or indicator organisms, antimicrobials and their metabolites, resistance genes, and mobile genetic elements. For these reasons the setting feeding into surveillance for antimicrobials includes not just hospitals or clinical microbiology labs, although they are important, but also animal health laboratories, watershed and soil monitoring programs, and routine animal health surveillance. Collecting and interpreting such varied data, then using it to inform public health programming, is challenging.
This chapter will first give a broad introduction to surveillance for antimicrobial resistance, explaining the types of systems collecting information and the way information moves through them, paying particular attention to various surveillance systems operating globally and the challenge of integrating data from these disparate sources. Next, it discusses the main types of relevant data collected and the inferences that can be made from them, with some attention to making better use of routinely collected clinical phenotype information and integrating this data into public health surveillance. The last section gives more attention to questions of monitoring resistance in water, sewage, and other environmental reservoirs, with the committee proposing steps to better characterize the relatively neglected environmental dimension of surveillance.
SURVEILLANCE SYSTEMS
Surveillance systems are critically important for understanding the burden of antimicrobial resistance, detecting the emergence and spread of resistant pathogens, targeting interventions to prevent and control the emergence of resistance, and measuring their effectiveness. Whether at the local, national, or international level, surveillance systems for antimicrobial resistance can differ considerably regarding their objectives, scope, and methods.
Surveillance systems can be passive or active. A passive system relies on self-reporting from organizations with relevant data, (e.g., health care facilities, animal health laboratories, or water monitoring programs) depending on the monitoring scope of the system. Passive surveillance is relatively inexpensive to manage because the labor costs to get the data are low, but the information gained is not likely to be complete, timely, or representative of the target population (Lee et al., 2010). In the context of antimicrobial resistance, active surveillance can involve the deployment of public health staff to monitor a target pathogen by actively contacting institutions and collecting information about the incidence of infections caused by that pathogen. Active surveillance can minimize problems with data completeness and representativeness and is available on a predictable timetable, but these advantages come at a cost. An example of an active surveillance is the Active Bacterial Core within the Emerging Infections Program of the Centers for Disease Control and Prevention (CDC), which has collected clinical information and resistance data for community-acquired infections of five invasive bacteria since the 1990s (Fridkin et al., 2015; GAO, 2020). The Active Bacterial Core surveillance program was able to identify important health disparities in risk of community-acquired methicillin-resistant Staphylococcus aureus (MRSA) in certain states, informing health policy and outreach decisions to counter this disparity (Fridkin et al., 2015).
Sentinel surveillance systems (active or passive) are based on surveillance of selected sites rather than being comprehensive across a location or population. These sites collect data from a large population in areas likely to find antimicrobial resistance hospitals or clinical laboratories, for example (Lee et al., 2010). The CDC’s Gonococcal Isolate Surveillance Program, which tracks antimicrobial resistance in gonococcal isolates submitted by 33 health departments across the United States, is an example of sentinel surveillance for resistant pathogens (CDC, 2021a).
Historically, surveillance for antimicrobial resistance has been built around human medicine, especially acute care. Data from acute care hospitals are often a starting point for surveillance as the clinical microbiology services in hospitals afford good quality data on the resistant pathogens causing infections in hospitalized patients. Newer tools for surveillance include genomic analysis that can be used to trace the source of infectious outbreaks, thereby contributing to a better understanding of the burden of antimicrobial resistance.
The design of a surveillance system to monitor antimicrobial resistance depends on decisions regarding the target to monitor, such as the microbial species, as well as the public health questions of interest. A concern with the effect of antimicrobial resistance on the environment might lead to a relatively greater interest in monitoring effluent (i.e., the discharging wastewater from treatment plants into the natural environment); an interest in characterizing the resistome of a mostly healthy population would lead to monitoring of a sample representative of this population,1 such as sewage. After considering such questions, there are also technical questions regarding how surveillance data will be shared, as well as sampling strategy and geographical scope. Box 4-1 gives an overview of some of these foundational questions as they relate to monitoring antimicrobial resistance.
Laboratory data reporting into surveillance systems can take several different forms. A labor-intensive approach is manual reporting with data entered via internet questionnaire forms. Manual file extraction followed by uploading to a central webpage or online database is another common method for reporting resistance data into surveillance systems. Both these approaches add to the workload for laboratory staff and health workers and to delays on data availability. A more efficient approach is direct, automated transmission from the laboratory information system to the surveillance system. This can facilitate real- or near real-time reporting, allowing for more useful analysis of emerging trends.
Real-time, cloud-based surveillance sometimes draws on proprietary data. In the United States, where automated testing for antimicrobial sus-
___________________
1 “All the genes [in a microbial community] that directly or indirectly contribute to resistance” make up the resistome (Wright, 2010).
ceptibility is the norm, medical device companies can access considerable information about resistance trends if the laboratories using their systems are willing to share their (anonymized) test results (Ruzante et al., 2021). A recent analysis of such data from 29 clinical laboratories using the BioFire gastrointestinal panel was able to monitor trends in acute gastrointestinal infection for almost 2 years, finding about 70 percent of infections caused by bacteria, especially Clostridioides difficile and enteropathogenic Escherichia coli (Ruzante et al., 2021). Such results should be interpreted with caution, as they were not obtained from a deliberate sampling frame designed to be representative of the population, but were nevertheless largely similar to CDC surveillance data in the relative rank and proportion of pathogens detected (Ruzante et al., 2021). It can also be difficult to interpret some of the results from the medical device company’s data, as patient confidentiality requires it be de-identified. Without data on patient age and medical history, for example, it is difficult to comment on the public health implications of the observed C. difficile colonization.
World Health Organization and Multilateral Support for Surveillance
In the United States and other high-income countries there are many ways to monitor indicators of antimicrobial resistance in humans, animals, and the environment. There are fewer options in low- and middle-income countries, partly because of constraints on laboratory capacity. Across settings, the most effective surveillance for antimicrobial resistance needs to
integrate confidential patient data with information from other sources, making an agreement for data management and confidentially of paramount importance (Seale et al., 2017). Procedures for integrating confidential data are set out in the World Health Organization’s (WHO’s) Global AMR Surveillance System (GLASS) manual (WHO, 2015). Implementing the systems outlined in the manual is beyond the capacity of many low-income countries, however (Seale et al., 2017). For these countries, a road map to surveillance of resistant pathogens sets out the data governance agreements required, as well as the choice of sentinel sites and coordinating laboratories, even arrangements for storing and transporting isolates and data management (Seale et al., 2017). This road map allows that automated testing systems may not be in place in these countries (Seale et al., 2017). Box 4-2 gives more background on the WHO GLASS program.
Recent work from the WHO indicates an interest in expanding GLASS to microbes from animal and environmental sources. This includes helping countries to increase their capacity to monitor, collect, and report data on resistance of extended-spectrum beta-lactamase-producing Escherichia coli in humans, poultry, and water bodies, including those containing wastewater from human and food animal sources (WHO, 2021c). In its implementation guidance for One Health surveillance in low- and middle-income countries, the WHO (2021c) recommended that the protocol be “simplified, integrated, [and] trans-sectoral.”
The WHO also collaborates with the Food and Agriculture Organization of the United Nations (FAO) and the World Organisation for Animal Health (known by the historical acronym OIE) to build a Tripartite Integrated Surveillance System on Antimicrobial Resistance and Antimicrobial Use (FAO and WHO, 2019). The Tripartite system envisions a global, web-based repository for resistance and uses data from human, animal, food, plant, and environmental sources; a request for proposals to develop such a platform was issued in March 2021 (WHO, 2021b). The Tripartite surveillance system is one of the four global projects supported by the United Nations (UN) AMR Multi-Partner Trust Fund, a pooled funding compact established in 2019 to support the Tripartite’s joint One Health effort to combat antimicrobial resistance (FAO et al., 2020; UN MPTF Office, 2020).
The WHO’s GLASS model of surveillance is a major step forward in monitoring antimicrobial resistance in low- and middle-income countries. The information gleaned from this network could be complemented by active surveillance, and there are numerous systems in place for active surveillance of antimicrobial resistance in low- and middle income countries (Ashley et al., 2018). As Figure 4-1 shows, global programs to fight malaria, tuberculosis, and HIV usually have a resistance-monitoring component; there is also considerable attention to surveillance in industry and academia.
Integrating Information from Disparate Sources
The challenge of effective surveillance for resistant pathogens comes in part from the useful information being collected by a wide and disparate group of systems. A recent review found that of 72 international surveillance networks developed since 2000, 34 were still active in 2018
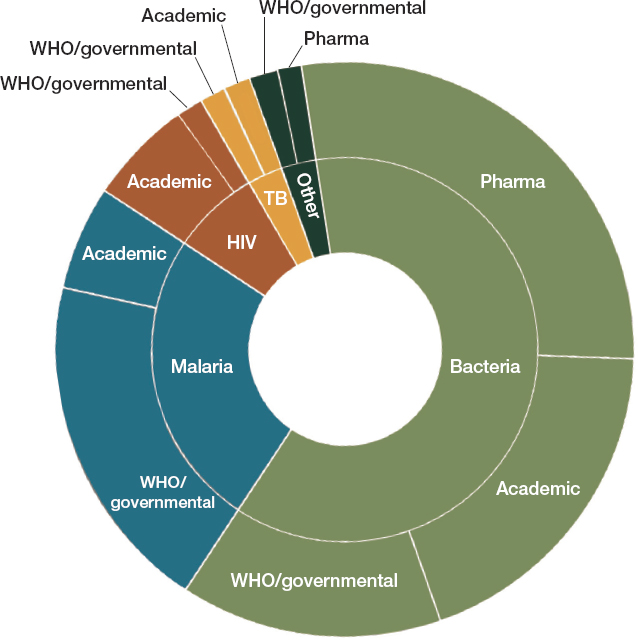
SOURCE: Ashley et al., 2018.
(Ashley et al., 2018). Of the 45 networks conducting surveillance for resistance in bacteria or fungi, 21 were still active in 2018 (Ashley et al., 2018). A more recent review identified 71 surveillance networks, mostly in Europe and the Americas, that monitored at least one species of resistant bacteria, though only 26 of these networks appear to be active (Diallo et al., 2020). Many of these were national networks, but others had a regional scope, such as the European Antimicrobial Resistance Surveillance Network and the Central Asian and Eastern European Surveillance of Antimicrobial Resistance.
More attention to monitoring resistance might yield more and better data to inform policy decisions, but it is not always so. Different systems collect data differently, and report results on different schedules, sometimes only once a year (Diallo et al., 2020). Only three of the active surveillance networks that Diallo and colleagues (2020) reviewed have
real-time monitoring with an alarm for the detection of critical pathogens. In general, different networks monitor animal and human health indicators. Some, such as the United States’ National Antimicrobial Resistance Monitoring System for Enteric Bacteria, look at both, but environmental monitoring is not a common component (Diallo et al., 2020). Harmonization efforts could do much to improve comparability of data and the speed at which it is shared (Diallo et al., 2020).
As Figure 4-1 shows, the private sector also operates surveillance networks. For example, GlaxoSmithKline has a regular survey of community-acquired respiratory tract infections in more than 30 countries; Merck has a program to look at antimicrobial resistance in isolates from intra-abdominal, blood stream, urinary tract, and respiratory tract infections in more than 60 countries (AMR Industry Alliance, 2017a,b; Enne et al., 2016; Hermsen et al., 2020; Torumkuney et al., 2016). The Pfizer Antimicrobial Testing Leadership and Surveillance database allows public access to both antifungal and antibiotic resistance data (Pfizer, 2021). Sometimes multiple companies collaborate in a surveillance consortium, as in the over 20-year SENTRY Antimicrobial Surveillance Program that collects and tests isolates from sentinel medical centers around the world (Fuhrmeister and Jones, 2019). Because of the large number of collaborators, isolates can be tested against 20 to 30 drugs, including investigational drugs (Fuhrmeister and Jones, 2019). This in turn informs antibiograms with scarce, valuable information about susceptibility to new drugs (a topic discussed more in Chapters 5 and 6) (Fuhrmeister and Jones, 2019).
In an effort to better understand private surveillance networks for antimicrobial resistance, the Wellcome Trust and the Open Data Institute established AMR Register, a clearinghouse for pharmaceutical companies’ human surveillance data (AMR Research Initiative, 2021a). In May 2021, Wellcome announced that it would work with the nonprofit organization Vivli to evolve the pilot project into a public website that publishes these data (AMR Research Initiative, 2021b; Vivli Center for Global Clinical Research Data, 2021).
Figure 4-1 also notes academic surveillance efforts. Some such programs are related to ongoing academic research projects, and as such have a set duration. Academic surveillance networks can also be run in collaboration with industry. The Global Point Prevalence Survey of Antimicrobial Consumption and Resistance, for example, relies on the sponsorship of bioMerieux, a diagnostics company, and the University of Antwerp to conduct point prevalence studies of antimicrobial consumption and resistance in over 800 hospitals in 80 countries (Biomerieux, 2020; Global PPS, 2021a,b). These surveys help in the evaluation of hospital stewardship programs, charting whether they are effective at reducing antimicrobial consumption or the emergence of resistance. Some academic networks are
wholly or partly devoted to low- or middle-income countries. The International Nosocomial Infection Control Consortium is one such network, with over 300 participating study sites in Latin America, Asia, and the Middle East and North Africa. (INICC, 2013). The consortium works to identify infection control strategies suitable to low- and middle-income countries; its surveillance component has helped quantify the higher risk of surgical site infections and device-acquired infections both three to five times higher in low- and middle-income countries than in high-income ones (Rosenthal, 2016).
These networks could provide a useful source of complementary data to inform estimates of the burden of the antimicrobial resistance. There is also ample room to make better use of, and have better access to, some of the data these networks collect, a topic discussed later in this chapter.
Automated Reporting
Automated surveillance systems have clear advantages over methods that rely on people to report data. Automation makes the reporting faster and easier, and it causes minimal disruptions to routine work. Nevertheless, a 2018 assessment of automated reporting from clinical diagnostic laboratories across Europe found that the most common barriers to automated reporting to the national surveillance systems were technological and financial (Leitmeyer et al., 2020). The laboratory software developers and vendors might not have developed ways to make their data compatible with the national surveillance systems as this is not their primary business (Leitmeyer et al., 2020). The protection of confidential information is a barrier to automated surveillance, especially in countries where there is no legal framework to support automated surveillance.
A lack of information technology is often cited as a barrier to comprehensive surveillance for antimicrobial resistance in low- and middle-income countries (Vong et al., 2017). Limited internet connectivity is a related challenge, as are the cost of the hardware, software, and staff to support automated surveillance networks (Vong et al., 2017). The WHO Collaborating Center for Surveillance of Antimicrobial Resistance has developed WHONET, a free, Windows-based software for microbiology laboratories to use for analysis of susceptibility test results (Vong et al., 2017). WHONET also provides a data conversion tool to ease the transfer of raw susceptibility data from the laboratory system to surveillance systems such as GLASS (Vong et al., 2017). Lab managers simply have to opt in to have the data uploaded to a surveillance system. Figure 4-2 illustrates the flow of information from hospitals or national sentinel sites to the national data hub, also highlighting some of the country support the WHO is providing for the surveillance of antimicrobial resistance.
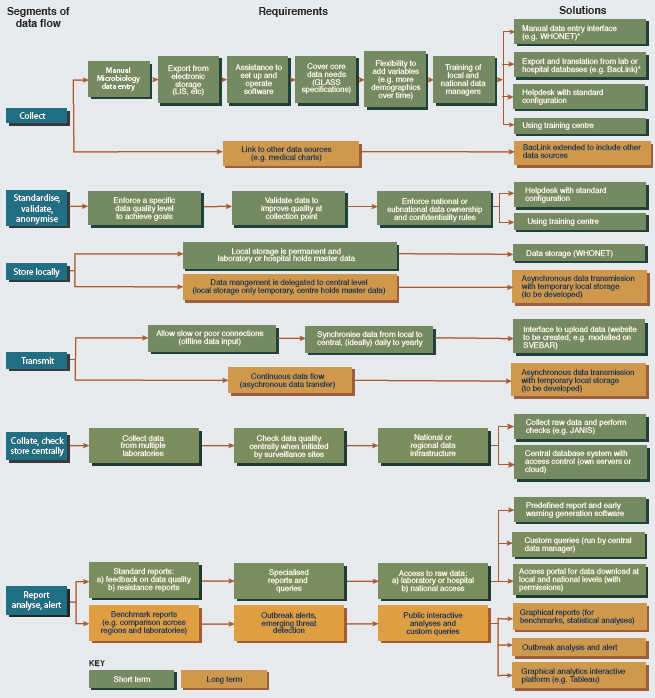
SOURCE: Reproduced from Using information technology to improve surveillance of antimicrobial resistance in South East Asia, Sirenda Vong, Amaury Anciaux, Anette Hulth, John Stelling, Visanu Thamlikitkul, Sunil Gupta, Jonas M. Fuks, Kāmini Walia, Pinyo Rattanumpawan, Sergey Eremin, Klara Tisocki, Tika R. Sedai, Anuj Sharma, Volume 358, 2017, with permission from BMJ Publishing Group Ltd.
FAO is working to adapt WHONET to collect antimicrobial resistance data on food and food-producing animals, with consideration to the elements of database design that allow for compatible automated data capture (FAO, 2019b, 2021a). To this end, the organization has developed a data management template and an online database from which information on antimicrobial resistance in food and animals will eventually be uploaded to the Tripartite surveillance system (FAO, 2019b, 2021b).
Such developments are promising, but in order for countries to participate in these nascent networks, they first have to have the capacity to
collect data. To this end, FAO provides assessment tools to measure capacity to monitor antimicrobial resistance in food systems and to set specific priorities for improvement (FAO, 2021a; FAO and UN, 2021). FAO also encourages regional cooperation to ensure harmonized data collection and regional comparability of results (FAO, 2019a). The WHO and the FAO’s Codex Alimentarius Commission have produced draft guidelines on monitoring foodborne antimicrobial resistance (FAO and WHO, 2021).
DATA COLLECTION AND ANALYSIS
Antibacterial resistance can be determined through two main sets of tools: phenotypic and genotypic. Phenotypic antimicrobial susceptibility tests determine the effectiveness of an antimicrobial compound in killing or inhibiting the growth of specific bacterial types. Such test results are vital for clinical decision making, including the drug and regimen for antimicrobial therapy. At a population level, susceptibility data are valuable indicators of trends in antimicrobial resistance (Cusack et al., 2019). There are also a wealth of genotypic tools, more widely used in research, that can help inform a better understanding of the burden of resistance. This includes multiplexed molecular panels incorporating resistance markers, which are becoming increasingly common in clinical and environmental laboratories and offer rapid and accurate genotypic susceptibility results and next-generation sequencing approaches.
Phenotypic Tests
The two most commonly used methods to test antimicrobial susceptibility are diffusion and microdilution. As Figures 4-3 and 4-4 show, both types of test rely on established clinical breakpoints to determine if an organism is resistant or susceptible to an antimicrobial compound or at an intermediate point between resistance and susceptibility, from which the microbiologist measures the minimum drug concentration needed to inhibit microbial growth (the minimum inhibitory concentration, or MIC). Of the two methods, broth microdilution provides more information on the extent of susceptibility or resistance that facilitates comparison of susceptibility profiles over space and time. The microtiter plate format is also more amenable to automation from setup to reading, as Figure 4-4 shows, although automated disc dispensers are commercially available to facilitate the setup of diffusion tests.
Both diffusion and microdilution tests are used in medicine to understand the clinical susceptibility of a pathogen to an antimicrobial. This is information a physician or veterinarian would need to identify a resistant infection and treat a patient. This clinical resistance is driven at the molecular level by traits in the pathogen that convey resistance to antimicrobials. Recent advances in testing have given better insight into the
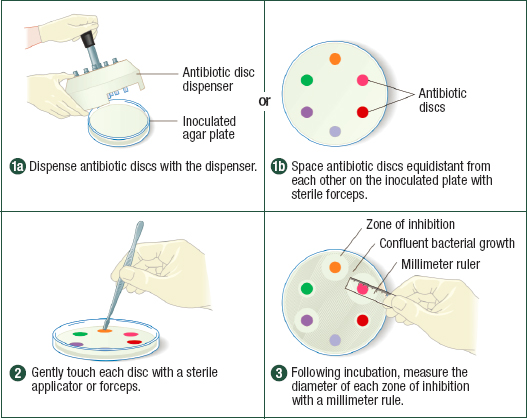
NOTES: In disk diffusion, different antimicrobials or different concentrations of the same antimicrobial are spaced on a culture plate inoculated with the pathogen of interest. After incubation, the diameter of the zone of inhibition around each disk is measured.
SOURCE: Cappuccino, James G. and Natalie Sherman; Microbiology: A Laboratory Manual, 10th Ed., ©2014. Reprinted by permission of Pearson Education, Inc.
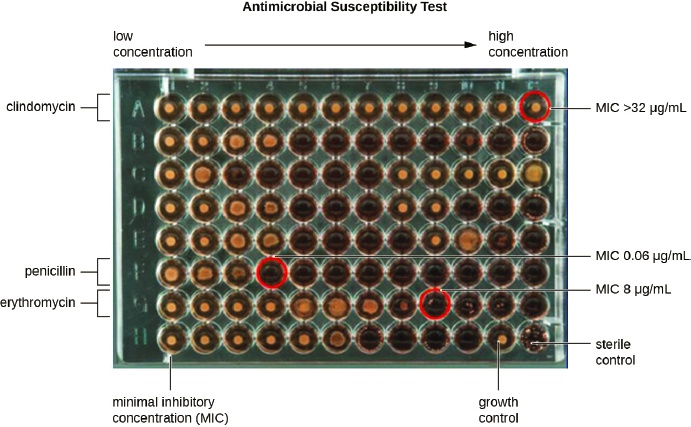
SOURCE: Lumen, 2021.
genetic basis of resistance, information that can be monitored in academic and public health surveillance efforts.
Genotypic Tests
Genotypic tools for identifying resistance look for sequences in the genetic code that indicate mechanisms of resistance to antimicrobials. Genotypic analyses include many tools based on molecular analysis, (i.e., polymerase chain reaction or PCR), which amplify sections of bacterial DNA to detect the presence of resistant traits. In clinical settings, molecular tests typically look for genes known to convey resistance to relevant drugs. The results can be helpful in informing clinical treatment by ruling out treatments for which the target pathogen carries resistance genes. At the same time, the presence of resistance genes does not necessarily predict treatment failure, nor does the absence of resistance genes necessarily indicate the clinical susceptibility of the pathogen, especially in gram-negative organisms (Bard and Lee, 2018; Galhano et al., 2021). Quantitative PCR (qPCR), also known as real-time PCR, not only detects the presence of a resistance gene but can also measure the concentration of the gene in the sample. For this reason, qPCR is frequently used to detect resistance genes in the environment. Its usefulness has increased with the introduction of high-throughput, real-time quantitative PCR, a technique that can analyze the presence and quantity of many resistance genes or mobile genetic elements at the same time, performing multiple assays using samples of only nanoliters (Franklin et al., 2021; Luby et al., 2016).
Whole genome sequencing identifies resistance by comparing the sequenced genome of the pathogen in question to known sequences that encode antimicrobial resistance (see Figure 4-5). Through the use of computerized algorithms and pattern recognition (called machine learning), genome sequences can be rapidly compared to their phenotypic resistance patterns to identify novel resistance mechanisms or mutations (Hendriksen et al., 2019a; Schurch and van Schaik, 2017).
Whole genome sequencing has been essential to the recent COVID-19 response, and using this tool in surveillance can be essential for outbreak response (Africa CDC, 2020) For this reason, a group of public, private, and nonprofit organizations, led by the African Union Commission through the Africa Centres for Disease Control and Prevention, recently invested $100 million to expand the use of genomic sequencing tools in public health surveillance and laboratory networks across Africa (Africa CDC, 2020).
There are also genotypic tools to describe the composition of microbial communities rather than single (cultured) organisms. As Figure 4-6 shows, metagenomic analysis can be especially useful in environmental monitoring (Schmieder and Edwards, 2012). Sequence-based metagenomic analysis involves extraction of DNA from an environmental sample and sequencing all or portions of it. The sequenced DNA—the metage-
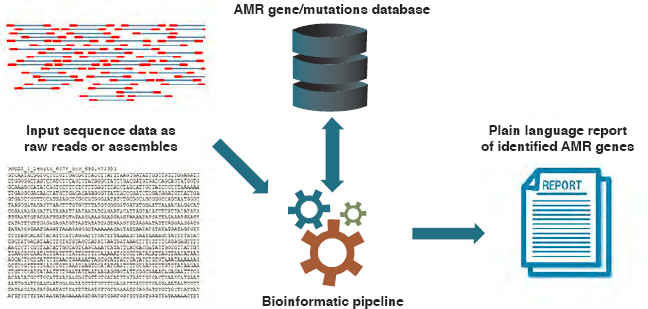
SOURCE: Hendriksen et al., 2019a.
nome—is then compared to a reference database of resistance genes and other determinants of resistance. (Examples of such reference databases include the Comprehensive Antibiotic Resistance Database, ResFiner, and the Reference Gene Catalog [Alcock et al., 2020; Bortolaia et al., 2020; Feldgarden et al., 2021a].) Functional metagenomics involves cloning the collective genome of all the organisms in a sample into a bacterial host, often E.coli, to identify resistance genes or genetic elements that may not be apparent from the sequence alone (Allen et al., 2010). This functional metagenomic analysis can be highly exploratory; no specific target pathogen, resistance gene, or mobile genetic element needs to be identified prior to the analysis. This can, therefore, be a very time-consuming approach to analysis, sometimes described as “tedious” or like searching for a needle in a haystack (Kowalchuk et al., 2007).
Genotypic tools are useful in surveillance: they are fast and require relatively small sample volumes. The ability to detect the presence of a resistance mechanism, even at low levels, is useful in surveillance. (The same is not true in clinical medicine, where the presence of resistance genes is not necessarily clinically useful information.) Genotypic tools are valuable in studying bacteria that are difficult to culture (Franklin et al., 2021). They have the potential to identify drivers of resistance, such as the presence of genes associated with resistance to heavy metals or multiple drugs or with mobile genetic elements (Franklin et al., 2021; Hendriksen et al., 2019a; Sundsfjord et al., 2004). They can also detect resistance mechanisms regardless of whether the pathogen is alive, a determination that does not rely on phenotypic breakpoints, which may be defined differently depending on the testing standard used (Sundsfjord et al., 2004).
At the same time, genotypic tests are currently expensive and require more sophisticated equipment and analysis than phenotypic susceptibil-
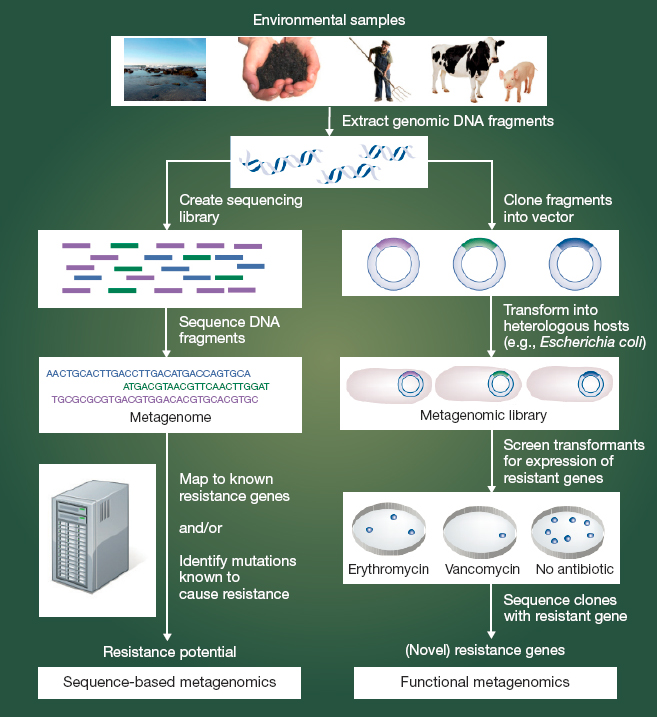
NOTES: Cloning fragments into an expression vector to create a metagenomic expression library is a new approach especially valuable in identifying genes involved in resistance to target antimicrobials. It is a complement to the more widely used metagenomic approaches delineated on the left.
SOURCE: Schmieder and Edwards, 2012.
ity tests (Wellcome Trust, 2018). What is more, resistance encoded by a previously unknown gene will be missed by genotypic methods based on sequencing. Furthermore, the presence of a resistance gene or determinant is not the same thing as its functional expression in a pathogen, something a phenotypic susceptibility test determines (Dunne et al., 2017). Looking at the RNA in a transcriptome can give insight into the genetically active and inactive components of a genome or microbiome (Shakya et al., 2019). The sequencing of RNA in microbial communities (called metatranscriptomics) can clarify the ways in which genes function (Franzosa et al., 2014;
Korry et al., 2020; Shakya et al., 2019). While metagenomic analysis gives insight into the structure and potential function of the microbial community, metatranscriptomic analysis gives insights into gene expression and function in response to specific conditions. Tools used together can provide a good understanding of changes in microbial communities and the functional response of antibiotic resistant genes in response to antibiotic exposure, if they can be adequately assembled and annotated from sequencing datasets (Korry et al., 2020). At the same time, obtaining intact RNA from an environmental sample is technically difficult and time consuming, challenges that should be weighed against their use (Maki et al., 2017).
Analysis of an environmental sample using PCR to search for particular resistance markers cannot determine the microbial host of the resistance gene, making it difficult to interpret if or how the resistance is conveyed to key pathogens (Luby et al., 2016). While the new high-throughput, real-time PCR techniques can test for multiple resistance genes at the same time, they are incapable of identifying novel resistance genes or emerging genetic elements (Franklin et al., 2021). There is also a need for standardization of procedures, especially for whole genome sequencing and metagenomic analysis (Hendriksen et al., 2019a). Table 4-1 summarizes advantages and disadvantages of different methods to identify the presence of antimicrobial resistance.
Identifying Resistance Patterns
Some systems in place for surveillance of antimicrobial resistance are designed to monitor resistance in particular pathogens or to a certain antimicrobial. AstraZeneca monitored susceptibility to meropenems in 21 countries for almost a decade, for example (Ashley et al., 2018). However, as protocols for phenotypic detection are streamlined and genotypic tools become cheaper and more accessible, coordinated monitoring across human, animal, and environmental samples will be more feasible.
One commonly used tool to monitor resistance patterns is the antibiogram, a “profile of antimicrobial susceptibility testing results of a specific microorganism to a battery of antimicrobial drugs” (Minnesota Department of Health, 2015). Antibiograms are usually presented in tables, pulling aggregate data from a hospital or health system (Minnesota Department of Health, 2015). Antibiograms are useful for monitoring trends in pathogens’ phenotypic resistance to different drugs; for this reason they are invaluable in both clinical medicine and surveillance. The production of antibiograms is part of the CDC’s Core Elements of Hospital Antibiotic Stewardship Programs (CDC, 2019a). Hospital reports, in turn, feed into state and county antibiograms that are used to both inform treatment decisions and monitor trends in resistance.
| Methods | Advantages | Disadvantages |
|---|---|---|
| Phenotypic tests | Low technical requirements Low cost per test Capable of identifying and quantifying antimicrobial-resistant bacteria Allows determining the phenotypic response of bacteria to selection pressure from antimicrobials Already in widespread use in clinical settings and water quality monitoring programs Ongoing global efforts to provide guidelines on collecting and reporting data |
Labor intensive and time consuming Inherent cultivation bias for fast-growing, easily cultivable bacteria Unculturable environmental bacteria that may serve as a reservoir of resistance are neglected For environmental surveillance, lack of benchmarking against culture-independent methods Results take days, necessitating prolonged use of broad-spectrum antimicrobials in clinical practice |
| qPCR and reverse transcriptase qPCR | Rapid quantification of target resistance genes for tracking, transport, and risk-assessment models With new high-throughput technologies, able to analyze a large suite of target genes simultaneously Ability to detect low-abundance genes (i.e., high sensitivity) Do not need live organism High specificity compared to many culture-based assays for environmental samples |
High technical requirements Inability to directly discriminate extracellular from intracellular DNA or the presence of resistance genes in live versus dead bacteria (qPCR only) In the case of reverse transcriptase qPCR, need for methods to avoid RNA degradation and preserve sample quality before processing Sensitivity can be influenced by qPCR inhibition Difficult to distinguish location of the gene (e.g., chromosome, plasmid, phage) Only amplifies a small region of the genome and may therefore detect pseudogenes (i.e., nonfunctional genes) |
| Whole genome sequencing | Antimicrobial-resistant bacteria can be typed and tracked by individual allele profile Achieves much higher resolution than traditional typing methods Determines co-carriage of specific genes causing different multidrug-resistance patterns |
Generally, cost prohibitive for large studies Limited to the individual bacterial cells that can be cultured and sequenced; newer technologies like single-cell genomics can capture unculturable microbes Requires accurate and up-to-date reference databases Very high technical requirements |
| Methods | Advantages | Disadvantages |
|---|---|---|
| Metagenomics | Able to analyze large numbers of relevant genes in environmental samples Can potentially carry out bacterial taxonomy and functional gene analysis simultaneously Using PCR-free library preparation removes problems with unsuitable primer design and PCR biases Can predict new variations on resistance genes Datasets can be compared between studies Helps to expand ARG databases |
Highest cost and technical requirements of all the methods Poor repeatability due to limitations in current analysis methods (in development); repeatability will increase with the development of guidelines and standards of analysis and the growth of curated reference databases Labor intensive with complex sample preparation and analysis Difficult to directly link the presence of a resistance gene with a specific resistant bacteria No live, dead, or active discrimination when not culturing first When PCR-dependent library preparation is used, PCR biases can affect analytical sensitivity and accuracy (e.g., exaggerations of dominant taxa or omitting low number abundance taxa) Does not provide enough sequencing depth to enrich and assemble genomes of a single strain (especially in complex matrices); however, this limitation depends on the platform Results dependent on library preparation and bioinformatics workflows |
| Metatranscriptomics | Allows the characterization and quantification of antimicrobial resistance genes that are metabolically active (being expressed) | Only characterizes resistance genes that are actively expressed at the particular time of sample collection and in that particular environmental condition Expensive Requires samples to be frozen immediately at ultra-low temperatures or stored in special preservatives |
SOURCES: Reprinted from Journal of Microbiological Methods, Vol 184, A. M. Franklin, N. E. Brinkman, M. A. Jahne, S. P. Keely, Twenty-first century molecular methods for analyzing antimicrobial resistance in surface waters to support One Health assessments, Copyright (2021), with permission from Elsevier. Based on Franklin et al., 2021; Korry et al., 2020.
Useful as they are, antibiograms do not typically give information into mechanisms of resistance, information that can be used to predict resistance patterns in microbe–drug combinations that are not part of the antibiogram (Sundsfjord et al., 2004). As Figure 4-5 showed, automated test panels have limited space, so the ability to make inferences about susceptibility in microbe–drug combinations that are not included in automated susceptibility tests is helpful. The use of whole genome sequencing to identify root causes of resistance is also useful when phenotypic resistance patterns change. In the Philippines, for example, the pairing of sequencing data with local antibiograms revealed that an increase in carbapenem resistance was attributable to horizontal gene transfer rather than the spread of a single resistant genetic clone (Argimon et al., 2020).
Whole genome sequencing is especially valuable in connection resistance patterns that emerge in different places or species or over a long time. It has been used in neonatal intensive care units to connect outbreaks of MRSA even when months pass between cases (Harris et al., 2013). Whole genome sequencing can also identify genetic links between resistant pathogens affecting humans and livestock (Davis et al., 2015).
The effectiveness of using phenotypic and genotypic data together to combat antimicrobial resistance can be seen in the Walter Reed Army Institute of Research’s Multidrug-Resistant Organism Repository and Surveillance Network (MRSN). Established to identify and prevent the spread of gram-negative multidrug-resistant organisms in military hospitals, the network provides a standardized system to interpret and compare resistance data across diverse settings (Chandrasekera et al., 2015). The MRSN also emphasizes prompt turnaround on testing to inform clinical practice. Pathogen identification and susceptibility are confirmed within 48 hours, along with PCR results screening for resistance genes (Chandrasekera et al., 2015). Information derived from whole genome sequencing and other advanced genomic tests are made available within a week (Chandrasekera et al., 2015). Through these efforts, MRSN supports attention to standard minimum data included on antibiograms, allowing for greater comparability of antibiograms across sites (Chandrasekera et al., 2015).
The MRSN allows clinicians prompt access to valuable information about resistant pathogens, informing treatment decisions and allowing them to put infection control measures in place quickly if necessary. It can also identify new resistant pathogens and genes. The value of the information gained is amplified through the Department of Defense’s (DOD’s) ongoing surveillance for resistant pathogens in host country military and civilian hospitals as part of the Global Emerging Infections Surveillance program (Health.mil, 2021; Meyer et al., 2011). Such monitoring can track the spread of antimicrobial resistance globally, informing clinical practice
and national policy for combating antimicrobial resistance (Meyer et al., 2011).
National Institutes of Health Efforts to Curate Information About Resistance
The MRSN may be unique in the population it serves and in the speed at which it is able to collect, analyze, and act on a signal, but it feeds into an even larger data collection effort. The National Center for Biotechnology Information (NCBI), a division of the National Library of Medicine (NLM) at the National Institutes of Health (NIH), has collected and made publicly available considerable information about resistance genes, genome sequences, antimicrobial susceptibility data, and bacterial genomes (NLM, 2019a). In 2016, it launched the Pathogen Detection Isolates Browser, an evolving tool that collects and analyzes gene sequences of pathogen isolates from human, food, animal, and environmental sources (NLM, 2019b). Submissions to the isolate browser come from a range of U.S. federal health agencies (including the DOD, the CDC, the Food and Drug Administration, and the U.S. Department of Agriculture [USDA]), state public health and agriculture laboratories, universities and hospitals, and international partners in the United Kingdom, Denmark, Australia, Canada, and Mexico (NLM, n.d.a). Figure 4-7 shows resources available in the Pathogen Detection Project and the relationships among them.
As Figure 4-7 indicates, public health and clinical laboratories can submit data to the NCBI databases, as can researchers or the managers of various data repositories (NLM, n.d.b).2 As of June 2021, the Pathogen Detection Isolates Browser contained nearly 900,000 isolates from 33 bacteria and the fungus Candida auris (NLM, 2021b). All data are publicly available.
The Pathogen Detection project has two objectives. The first is to identify relationships among disparate clones as part of the source investigation and response to on an outbreak (NLM, n.d.a). The other is to facilitate monitoring and research on resistance genes and other relevant genetic information that encode antimicrobial resistance. To this end, NCBI has built the National Database of Antibiotic Resistant Organisms (NDARO) (NLM, n.d.c). This database includes a reference catalog with the capability to search for acquired resistance genes and proteins and point mutations as well as genes encoding for co-resistances (including metal and biocides) and virulence (Sayers et al., 2021). It includes software that analyzes all uploaded isolates, with the exception of C. auris, for
___________________
2 Submission requires a record of the project in NCBI’s BioProject database, a record in NCBI’s BioSample database with the isolate metadata for each pathogen sequenced, and a submission of the raw sequence data to NCBI’s Sequence Read Archive database (NLM, 2021a).
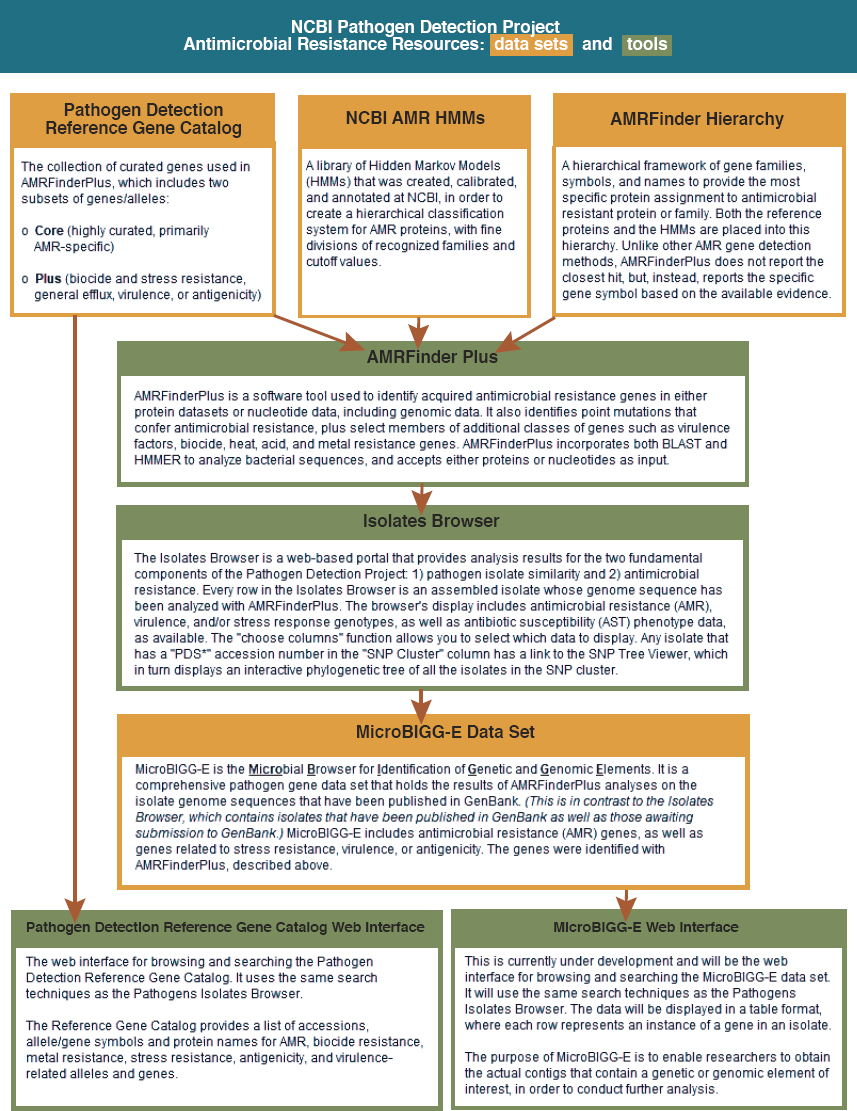
SOURCE: NLM, 2020.
resistance genes identified in the databases, with the capacity to identify co-resistance and point mutations for some pathogens (Feldgarden et al., 2021a). Antibiogram data from phenotypic susceptibility tests can be uploaded and stored with the sequence. The phenotypic information is included as a column in the Isolate Browser next to the column of identified resistance genes, as well as details of the antibiogram, including minimum inhibitory concentrations and the testing standard used. Additional information about the isolates (e.g., clinical or environmental
sample; from blood, urine, meat; from retail or wholesale) is included in the metadata. The database can be searched for combinations of phenotypic and genotypic resistances, including co-resistances and virulence. As of January 2021, the project has included the Microbial Browser for Identification of Genetic and Genomic Elements, which allows users to download protein and nucleotide sequences as well as isolate metadata about assumed phenotypic expression (Sayers et al., 2021).
The committee commends NCBI for its Pathogen Detection and NDARO programs, both for scope of the programs and the breadths of contributors they are accessible to. These databases are not only invaluable tools for researchers and public health investigators, but they are useful to inform discussion about priorities for medicine and diagnostic development.
However, the focus of the programs on genotypic data leaves much available data on resistance unmined. Of the almost 900,000 isolates submitted to the browser, only a little more than 10,000 entries (about 1 percent) have associated antibiogram data (Hendriksen et al., 2019a; Prasad, 2021). Most of these antibiograms have been entered by federal agencies, and more than half are for Salmonella spp. (Strain, 2021). Antibiogram data from many hospitals, universities, and public health laboratories around the country are not being uploaded. The committee encourages NCBI to continue its efforts to communicate about the Pathogen Detection Project to all parties who may have data to contribute, emphasizing the importance of including antibiogram data when available.
As seen with the example of MRSN, collecting phenotypic data within a broader database can facilitate the aggregation of antibiograms and bolster efforts to depict the emergence and spread of resistance (CDDEP, 2021). The sequencing of resistance genes and associated mechanisms of resistance can increasingly predict pathogens’ susceptibility to different antimicrobials (Hendriksen et al., 2019a; Schurch and van Schaik, 2017) (see Table 4-2). It is still too expensive and resource intensive to be useful in clinical practice, especially in low- and middle-income countries, however (NIHR Global Health Research Unit on Genomic Surveillance of AMR, 2020). For surveillance purposes, genome sequences may be sufficiently reliable for identifying patterns of antimicrobial resistance of public health importance (Bortolaia et al., 2020; Koser et al., 2014; Schurch and van Schaik, 2017). As the GLASS protocols acknowledge, phenotypic testing is the common denominator of global surveillance (Bard and Lee, 2018).
A Phenotypic Database
Phenotypic susceptibility tests are a mainstay of clinical microbiology. They are also valuable in environmental surveillance as they are relatively
inexpensive and allow for comparison with other epidemiological data. Some water monitoring programs already rely on phenotypic susceptibility tests (Bard and Lee, 2018; Franklin et al., 2021; McLain et al., 2016). There is a great deal of susceptibility data generated daily at U.S. hospitals, nursing homes, diagnostic laboratories, and environmental monitoring sites on phenotypic resistance that is not captured by the Pathogen Detection Project because no genomic data are collected to which the phenotypic results can be attached. Even if there are genomic data to which susceptibility results can be linked, adding this information creates an extra manual step on the part of the submitter. Furthermore, as evidenced by the efforts of the Wellcome Trust and the Open Data Institute to establish the AMR Register, there is considerable relevant industry data that is not yet public and therefore not captured in the NCBI databases. The phenotypic information from these sources would be useful for understanding the prevalence, spread, and evolution of resistance. A reasonable approach to incorporating genomic data is to monitor markers of genomic resistance alongside phenotypic susceptibility data when available. Resistance detection assays, as part of multiplexed molecular panels including syndromic panels, have increased the availability of such results that may also inform treatment decisions (Dien Bard and McElvania, 2020).
The committee recognizes the rapid advancements that have been made in genome sequencing. Genomic tools are increasingly used to identify patterns in antimicrobial resistance and predict susceptibility to antimicrobials. Chapter 6 discusses how such tools can be used to inform drug discovery. The wealth of research tools presented in Table 4-1 are all used, individually and together, to better understand the way resistant pathogens move through human and animal hosts and the environment. Rapid sequencing can help detect emerging pathogens passing between humans and animals (GAO, 2021). The USDA and the CDC use these tools to track emerging variants of SARS-CoV-2 in humans and animals (APHIS, 2021; GAO, 2021). Similarly, these tools drive forward research on antimicrobial resistance and efforts to counter it.
Excitement over the immense potential of new analytic technologies should not blind us to the value of the vast amounts of information on phenotypic antimicrobial susceptibility currently generated in clinical laboratories and health departments all over the world. There is useful data in ordinary antibiograms that could inform a better understanding of the trends in antimicrobial resistance. Failure to make full use of this data for understanding disease burden and emerging trends is wasteful.
Phenotypic data are useful for surveillance, even in the absence of corresponding genotypic information. Furthermore, the amount of phenotypic data generated regularly in clinical laboratories, both from animal and human samples, dwarfs that being produced via whole genome
| Pathogen | Number of Pathogens | AST Method | Number of Antimicrobials | Bioinformatic Tool | Sequencing Data | |
|---|---|---|---|---|---|---|
| 2013 | S. Typhimurium E. coli E. faecalis E. faecium |
49 48 50 50 |
MIC | 17 14 |
ResFinder | Assembled, Velvet |
| 2013 | E. coli (ESBL) K. pnuemonia (ESBL) |
74 69 |
DD | 7 | BLASTn, selected panel | Assembled, Velvet |
| 2014 | S. aureus | 501 | DD/MIC (Vitek) | 12 | BLASTn, selected panel | Assembled, Velvet |
| 2016 | C. jejuni C. coli | 32 82 | MIC | 9 | BLASTx | Assembled, CLC-bio |
| 2016 | S. enterica | 104 536 |
MIC | 14 | ResFinder/ARG-ANNOT/CARD/BLAST | Assembled, CLC-bio |
| 2017 | E. coli K. pnuemonia P. aeruginosa E. cloacae |
31 24 22 13 |
MIC | 4 | Custom DB based on ARDB/CARD/ß lactamase alleles | |
| 2017 | S. enterica E. coli C. jejuni |
50 50 50 |
MIC | 4 6 4 |
ResFinder/PointFinder | Assembled, SPAdes |
| 2018 | E. faecalis E. faecium |
97 100 |
MIC | 11 | ResFinder/NCBI Pathogen DB/BLAST | Assembled, CLC-bio |
| 2018 | S. aureus | 501 491 397 |
DD/MIC | 12 | GeneFinder/Mykrobe/Typewriter | FASTQ/assembled, BLAST |
| 2018 | M. tuberculosis | 10209 | MGIT 960 |
4 4 4 4 |
Cortex | Assembled |
| 2019 | H. pylori | 140 | MIC (E-test) | 5 | ARIBA | FASTQ |
| Pathogen | Concordance | Sensitivity | Specificity | Comment | |
|---|---|---|---|---|---|
| 2013 | S. Typhimurium E. coli E. faecalis E. faecium |
99.74% | Disagreement: 7 isolates including 6 E. coli resistant to SPEC | ||
| 2013 | E. coli (ESBL) K. pnuemonia (ESBL) |
96% | 97% | VM rate: 1.2% M rate: 2.1% |
|
| 2014 | S. aureus | 97% | 99% | VM rate: 0.5% M rate: 0.7% |
|
| 2016 | C. jejuni C. coli | 99.2% | Lower accordance to GEN, AZI, CLIN, TEL | ||
| 2016 | S. enterica | 99.0% | 99.2% 97.6% |
99.3% 98% |
Lower accordance to aminoglycosides/ß-lactams |
| 2017 | E. coli K. pnuemonia P. aeruginosa E. cloacae |
87% | 98% | Negative predictive value: 97% Positive predictive value: 91% |
|
| 2017 | S. enterica E. coli C. jejuni |
98.4% | Disagreement: 2/2 C. jejuni to FQ/ERY 5 E. coli to Col (pmrB) |
||
| 2018 | E. faecalis E. faecium |
96.5% | |||
| 2018 | S. aureus | 98.3% | Disagreements: 0.7% Predicted resistant 0.6% Predicted susceptible |
||
| 2018 | M. tuberculosis | 89.5% | 97.1%/99.0% predicted R/S 97.5%/98.8% predicted R/S 94.6%/93.6% predicted R/S 93.1%/96.8% predicted R/S |
||
| 2019 | H. pylori | 99.% | Phenotype issues to metronidazole |
(1) ESBL: Extended Spectrum Beta-Lactamase, (2) MIC: Minimum Inhibitory Concentration, (3) DD: Disk diffusion, (4) VM: Very Major, (5) M: Major, (6) R/S: Resistant/Susceptible, (7) SPEC: Spectinomycin, (8) GEN: Gentamicin, (9) AZI: Azithromycin, (10) CLIN: Clindamycin, (11) TEL: Telithromycin, (12) FQ: Floroquinolone, (13) ERi: Erithromycin, (14) COL: Colistin
SOURCE: Hendriksen et al., 2019a.
sequencing and metagenomics. Central collection and analysis of phenotypic data would give better insight into the regional and global distribution of resistant pathogens than genomic results from the smaller subset of sequenced isolates. It could also provide a more representative picture of disease burden, as most investigators would only sequence pathogens that seem out of the ordinary.
Recommendation 4-1: The National Library of Medicine (NLM) should establish an open-source, unified antimicrobial resistance database that integrates raw phenotypic data from national and international efforts. This database should support automatic importation from hospitals, laboratories, and surveillance networks and linking to genotypic data when available. NLM should engage the Centers for Disease Control and Prevention, the U.S. Department of Agriculture, and other relevant stakeholders to determine the necessary data elements and confidentiality procedures.
An automated data ingestion pipeline would mitigate the additional burden on clinical laboratories imposed by such a database by taking disparate formats of collected antimicrobial resistance data and, either by a simple set of translation rules or potentially using more advanced machine learning techniques, automatically format and deposit the data in a consistent fashion. As commonly used automatic testing machines, including the bioMérieux Vitek and BD Phoenix, are already internet connected, once an initial pipeline from the laboratory information system is established, data deposition from these devices could be almost fully automated with laboratory intervention only in the case of errors. A standard data deposition form could also help new laboratories or regional laboratories that currently do not generate their own standardized antibiograms by providing a default template, contributing to more standardized measurements across the United States and internationally.
Automating Data Deposition
The WHONET software could be useful to diagnostic laboratories looking to automate data deposition, the process by which data are consolidated for analysis. WHONET does not require adoption of any specific laboratory information system, which can be costly (WHONET, 2020). For laboratories that already use such information management systems, WHONET has a free data importing tool (BacLink) to capture and standardize existing data (AHRQ, 2014; WHONET, 2021). Given its convenience and ease of use, over 2,300 laboratories (including human and
animal clinical laboratories, public health, and food safety laboratories) in 130 countries use WHONET, including for generating antibiograms (AHRQ, 2014; WHONET, 2021). By simply granting a centralized database permission to extract their data from WHONET, lab managers could contribute raw susceptibility data to a centralized database without any additional labor, much as is done for uploading data to GLASS. FAO is currently using WHONET tools for surveillance in food and food-producing animals. Further expansion of WHONET into animal health and environmental monitoring labs could ease automated data collection that the proposed database would draw from. The committee envisions the proposed database would pull phenotypic data from various human, animal, and environmental sources. Private-sector information, such as that the nascent AMR Register is collecting, would further enrich this repository, as would submissions from academic researchers.
Database Development and Use
One of NLM’s main concerns with expanding antimicrobial resistance data collection will be protecting privacy. For example, hospitals may not want antibiograms identifiable for fear of bad publicity or being labeled as a resistance hotspot. Some care must be taken to make the database anonymized while still useful. The use of WHONET is also relevant to this concern as software automatically removes identifiers from raw data (WHONET, 2020).
NLM has experience in the technical work of database development and familiarity with common stumbling blocks such as insufficient attention to controlling the reporting burden or removing data identifiers. NLM has already given attention to how to collect this type of information in the Pathogen Detection Isolate Browser discussed earlier, providing precedent for some important variables to collect data on (NCBI, 2019). Key variables found in data collection for the Isolate Browser that would also be collected in the proposed phenotypic database include pathogen species, the type of isolate (e.g., clinical or environmental), its source (differentiating, for example, between human and animal isolates, and among them isolates from blood, urine, tissue), and collection date (NCBI, 2019). Other optional, but useful, information would include where the sample was collected and patient diagnostic information if relevant and available.
The Pathogen Detection Isolate Browser also has a field for susceptibility test phenotypes, listing all the medicines tested against the isolate (NCBI, 2019). However, this field is currently limited to the categorical interpretation that the pathogen is resistant, intermediate, or susceptible to a drug (or “other” for pathogen–drug combinations for which breakpoints are not established). In the browser’s current capabilities, mini-
mum inhibitory concentrations are available if an antibiogram is attached to the genomic data (NLM, 2018). The committee notes that the usefulness of this tool and its comparability to other databases would be improved by the inclusion of numerical minimum inhibitory concentrations.
In a unified antimicrobial resistance database, numerical measurements or ranges of minimum inhibitory concentrations would be a crucial data field. Simply knowing that a pathogen is sensitive to a treatment is sufficient to guide therapy, but some susceptible pathogens are much closer to developing resistance than others. At the riskier end of the spectrum are sensitive pathogens that are only a few mutations away from resistance; such pathogens are of concern for public health surveillance. Increasing minimum inhibitory concentrations can provide forewarning of pathogens that may develop resistance (Baquero, 2001). Furthermore, the categorical cutoff points of susceptible, intermediate, and resistant are not fully standardized by pathogen, drug, or region, making comparisons difficult. Access to the minimum inhibitory concentrations would also allow researchers to reinterpret data as susceptibility criteria change (McLain et al., 2016). The proposed database would also be equipped to capture zone diameter measurements for data produced from manual susceptibility tests such as disk diffusion, discussed more in Chapter 6.
More information about minimum inhibitory concentrations could also inform determinations of epidemiological cutoff values (Martinez et al., 2015; McLain et al., 2016). Including phenotypic results from environmental monitoring sites increases the amount of data available to support this determination. As of June 2021, more than 62 percent of the sequenced isolates in the Pathogen Detection Project were from clinical settings (Feldgarden et al., 2021b). Efforts to diversify the sources of information will capture a more accurate, One Health picture of the true burden of resistance (Berendonk et al., 2015; McLain et al., 2016).
Finally, it would be helpful for the proposed database to capture information about genotypic resistance markers if available. Some resistance patterns are commonly monitored in clinical settings, including mecA, a gene associated with methicillin-resistant Staphylococcus aureus (MRSA) and van A and van B, conferring vancomycin resistance (CDC, 2019b; Saadat et al., 2014). Tests for extended-spectrum beta-lactamases (ESBL), enzymes that can break down the beta-lactam family of antibacterials, and carbapenemase-producing organisms, which can break down carbapenems, are also sometimes available and would be useful optional data to include. It could also be helpful to know, for clinical specimens, whether the infection was acquired in a hospital or in the community, allowing that this distinction is not always clear.
In the implementation of this recommendation, NLM would work with the CDC and the USDA as well as other relevant industry and
academic stakeholders to further discuss what data would be essential and what would be optional. This cooperation would also ensure that the database was set up to be most useful to the agencies responsible for surveillance and to set up a system easily mined by researchers from different disciplines. It would also be important to clarify a communication strategy or mix of incentives to encourage people to submit their data. The relevant professional societies for microbiology, including the American Society for Microbiology, the American Society for Clinical Pathology, and the Association for Molecular Pathology, would be valuable stakeholders to involve in any discussion of incentives for participation. The societies could help draw attention to the program and support laboratory staff to implement it.
The committee recognizes the value of contributions to surveillance of antimicrobial resistance already undertaken by the CDC; the USDA’s National Agricultural Research and Monitoring Surveys (NARMS), discussed later in this chapter, give similar insight into resistance patterns in agriculture. At the same time, there is considerable information generated outside of these networks, much of it having little life beyond the immediate purpose it was generated for. Hospital antibiograms, for example, may inform some mandatory reporting to county or state health authorities but are otherwise not used beyond the hospital.
Over time, the implementation of this recommendation could also allow for greater representation of surveillance data from low- and middle-income countries. Automated susceptibility testing is less common in low- and middle-income countries, partly because of the expense of the equipment and unreliable distribution systems for the consumables needed to operate them (Iskandar et al., 2021; Pascucci et al., 2021). However, most large hospitals in low- and middle-income countries do produce antibiograms (Iskandar et al., 2021; Pascucci et al., 2021; Tiwari et al., 2009). The Clinical Laboratory Standards Institute has guidance on how to standardize antibiograms, making them more accessible in places that rely on manual testing (CLSI, 2014). There is also growing interest in the use of widely accessible technology such as smart phones to generate antibiograms in settings with few resources (Pascucci et al., 2021). Eventually this could mean more phenotypic data to characterize antimicrobial resistance in parts of the world where the problem is worst, and potentially more antibiogram data to contribute to international surveillance efforts.
MONITORING ANTIMICROBIAL RESISTANCE IN WATER
Increasing the availability of information about environmental isolates collected by surveillance networks and stored in the proposed NLM database would contribute to a more holistic understanding of antimicro-
bial resistance. However, environmental monitoring of resistance is still new. Most environmental bacteria cannot be cultured, so until the recent advent of genomic tools, they could not be easily assessed for resistance mechanisms (Allen et al., 2010). As the field is still developing, methods for evaluating resistance in environmental isolates are not standardized (Berendonk et al., 2015; Murray et al., 2021; Pruden et al., 2018). As discussed in Chapters 1 and 2, antimicrobial resistance exists naturally in the environment. It is difficult to distinguish background levels of resistance from that caused by humans (Rothrock et al., 2016). The geographic scale of the area to be monitored adds to the challenge, as do the numerous targets for surveillance of antimicrobial resistance (e.g., pathogens, resistance genes, antimicrobials and residues, mobile genetic elements).
Furthermore, the number of antimicrobial resistance genes isolated from human and animal pathogens is much greater than what has been described in environmental bacteria (Berendonk et al., 2015). Some of the metagenomic data from environmental samples are thought to be resistance genes in the sample, but this characterization is often based on genetic similarity to resistance genes described in clinical samples, not on a functional demonstration (Berendonk et al., 2015). The Comprehensive Antibiotic Resistance Database is unique in that it only includes genes that have been characterized clinically or experimentally (Alcock et al., 2020). In other public databases, the description of resistance genes is more frequently putative (Berendonk et al., 2015).
The evolution of resistance in the environment and its transmission to humans is a serious concern, and the first step to improve understanding of that risk is monitoring resistant pathogens, resistances genes, and related genetic elements in the environment (Manaia, 2017). The mechanisms through which resistance moves from the environment to humans and other animals are not clear, and it is difficult to demonstrate if and how resistance traits in the environment influence clinical presentation in humans or other animals (Vaz-Moreira et al., 2014). Determinants of resistance in soil, surface water, and groundwater are thought to contribute to a reservoir of antimicrobial resistance in the environment (Dantas et al., 2008). Though the concentration of antimicrobials in water is generally low, it may still be enough to encourage the emergence of resistance (Murray et al., 2021). The challenge for environmental monitoring is to determine what factors amplify resistance genes in the environment and what factors encourage their transmission.
Monitoring sewage can provide access to samples from a large and diverse population, reflecting exposure to pathogens, resistant organisms, resistance genes, antimicrobials and residues, and heavy metals. Unlike surveillance methods that rely on clinical laboratory data, surveillance of wastewater gives insight into a largely healthy population and can capture
a wider range of relevant bacteria and resistance traits; sewage can sample, in essence, an entire city at one time (Aarestrup and Woolhouse, 2020; Brinch and Aarestrup, 2020). For this reason, the Global Sewage Surveillance project has been monitoring markers of antimicrobial resistance in sewage in 60 countries since 2016 (Aarestrup and Woolhouse, 2020; Brinch and Aarestrup, 2020; Hendriksen et al., 2019b). This project’s metagenomic analysis of sewage allows insight into the composition of the resistome, all the resistance genes in a bacterial community, including those of clinical interest as well as those in nonpathogenic bacteria that are not as well studied (Hendriksen et al., 2019b; Wright, 2010). This research has established geographic clustering of resistance patterns (Hendriksen et al., 2019b). In low- and middle-income countries metagenomic analysis of sewage samples has shown the relative abundance of resistance genes expressed are broadly consistent with patterns of antimicrobial use (i.e., genes that convey resistance to macrolide antibacterials are more abundant in places where macrolides are more commonly used) (Hendriksen et al., 2019b).
Box 4-3 describes how the COVID-19 pandemic has brought renewed attention to monitoring wastewater for infectious disease surveillance. It is possible that the pandemic will hasten use of wastewater for infectious disease monitoring more broadly. Monitoring sewage discharged from hospitals, for example, could give insight into the burden of both antimicrobial residues and pathogens in a point of likely antimicrobial pollution. Hospital sewage has been implicated in the dissemination of resistance, though the extent to which this risk is diluted at wastewater treatment is not clear (Buelow et al., 2020). Research on how the indicators of antimicrobial pollution move through the water supply, from hospital or factory effluent through wastewater treatment, through the environment and into the community would be invaluable for both characterizing this risk and identifying points where additional monitoring would be valuable. The comparison of resistance indicators at different points in the wastewater supply could identify resistance trends as they emerge, including during the treatment process. Research on indicators of resistance in water and the effect of sewage and wastewater treatment and their downstream activity could also inform mitigating actions in water policy.
Additional research could clarify how or where to monitor antimicrobial residues to inform environmental surveillance (Polianciuc et al., 2020). As Figure 4-8 shows, antimicrobial medicines and resistant pathogens can both enter the water supply through many paths. Industrial effluent, including factory effluent from pharmaceutical plants, is one avenue, as are agricultural runoff and human sewage (Fouz et al., 2020; Murray et al., 2021). Concentrations of antimicrobials in industrial waste are generally highest, about a thousand times higher than in wastewater, and a million times higher than in surface water (Murray et al., 2021). The
concentrations that select for resistant bacteria in water are difficult to establish, partly because the aquatic microbial community is so complex, with many antimicrobials and resistance genes mixing, as well larger forces—such as competition for nutrients—at work (Bengtsson-Palme and Larsson, 2016). Recent research efforts to predict antimicrobial concentrations at which the environmental resistomes would be unaffected are a valuable starting point for ecological testing and environmental risk assessment (Bengtsson-Palme and Larsson, 2016).
Environmental risk assessment is required in the United States and Europe when medicines are predicted to be found in a concentration in water above a certain threshold, but most environmental toxicological testing does not target bacteria, and the current thresholds might not protect
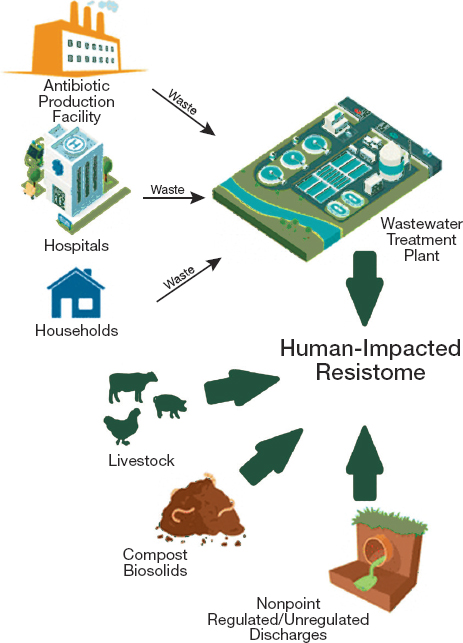
NOTES: Small arrows reflect waste flows from environmental reservoirs to a wastewater treatment plant. Large arrows reflect direct effects of different reservoirs on the human-affected resistome.
SOURCE: Reprinted and adapted with permission from Vikesland, P. J., A. Pruden, P. J. J. Alvarez, D. Aga, H. Burgmann, X. D. Li, C. M. Manaia, I. Nambi, K. Wigginton, T. Zhang, and Y. G. Zhu. 2017. Toward a comprehensive strategy to mitigate dissemination of environmental sources of antibiotic resistance. Environmental Science & Technology 51(22):13061-13069. ©2017, American Chemical Society.
against the emergence of antimicrobial resistance (Murray et al., 2021). What is more, this concentration is difficult to predict (Bengtsson-Palme and Larsson, 2016). Some evidence suggests that the minimum concentration at which an antimicrobial can select for resistance in microbial communities in water can be up to 200 times lower than the clinically meaningful minimum inhibitory concentrations (Murray et al., 2021). It is difficult to predict minimum selective concentrations because of a dynamic effect any one antimicrobial residue can have on a microbial community, and because of the interplay different medicines residues would have
with each other and with other stressors in the environment (Pruden et al., 2018).
Lack of consensus over what indicators of resistance to monitor in the environment holds back the ability of policy makers to monitor it. Some researchers have suggested the need for a composite antimicrobial contamination measure that could help identify environmental hotspots—nutrient-rich environments with high concentrations of bacteria (Pruden et al., 2018; Vikesland et al., 2017). Such an indicator could help distinguish the pathogenic resistance driven by humans from background level resistance that may not be a meaningful threat to health. One standard that could be used to narrow resistance genes of possible threat to human health is the likelihood that a resistance gene could be acquired and expressed in human pathogens (Martinez et al., 2015). Genes that are known to confer resistance in bacteria and are found in mobile genetic elements are of particular concern, especially if they are found in environments closely associated with people (Martinez et al., 2015). Berendonk and colleagues proposed 16 genetic determinants to use as possible indicators based on these criteria;3 other genes have been added more recently. The authors also suggested six bacterial groups as priorities for water monitoring efforts because of their likelihood to carry resistance genes or acquire them from environmental sources,4 their usefulness as indicators of water quality, and their frequency in both animal gut and environmental samples (Berendonk et al., 2015).
U.S. Government Work to Monitor Antimicrobial Resistance in Water
The 2021–2025 strategic plan for the National Antimicrobial Resistance Monitoring System includes a pilot project to monitor surface water for evidence of antimicrobial resistance (Garland, 2020). The project will also set out a standardized system for sampling and analysis of evidence of resistance in surface water (Garland, 2020). The program’s choice of surface water to monitor stems from the constant human exposure to surface water both directly through drinking, swimming, or other rec-
___________________
3intI1 (integrase gene of class 1 integrons, a genetic platform for resistance gene capture), sul1 and sul2 (sulfonamide-resistant dihydropteroate synthase), blaCTXM and blaTEM (beta-lactamases, frequently identified in Enterobacteriaceae), blaNDM-1 (New Delhi metallo-beta-lactamase), blaVIM (carbapenemase, frequent in clinical Pseudomonas aeruginosa in certain areas), blaKPC (Klebsiella pneumoniae carbapenemase), qnrS (quinolone pentapeptide repeat family), aac(6´)Ibcr (aminoglycoside acetyltransferase), vanA (vancomycin resistance operon gene), mecA (penicillin-binding protein), ermB and ermF (rRNA adenine N-6methyltransferase, associated with macrolide resistance), tetM (ribosomal protection protein, associated with tetracycline resistance), and aph (aminoglycoside phosphotransferase).
4Escherichia coli, Klebsiella pneumonia, Aeromonas spp., Pseudomonas aeruginosa, Enterococcus faecalis, and Enterococcus faecium.
reational use, and indirectly as with exposure of food crops to irrigation water (Franklin et al., 2021). Surface water is also an attractive candidate for monitoring because it is a natural endpoint and mixing site for treated wastewater, agricultural runoff, and other nonpoint water sources.
The Environmental Protection Agency (EPA) is in the process of analyzing samples collected from national surveys for the presence of six resistance genes and the mobile genetic element marker intI1 (most of them among the Berendonk and colleagues’ proposed indicators) and fecal indicators (Berendonk et al., 2015; Garland et al., 2018). Preliminary analyses indicate high concentrations of intlI1 with apparent hotspots in the Northeast and northern Midwest, sul1 with hotspots in the Northeast and central states, and tetW with hotspots mostly across the northern central states (Garland et al., 2018). Continued monitoring will give better insight into where and how the selected resistance indicators emerge and change over time.
The CDC also has pilot programs looking at resistant bacteria in surface water. E. coli is a commonly monitored indicator of water quality, and those E. coli that produce ESBL are a serious threat to human health (FDA, 2021). After a year of monitoring, CDC researchers found that about 70 percent of samples contained detectable ESBL-producing E.coli, with 84 percent of isolates resistant to two or more classes of antimicrobials (FDA, 2021).
Monitoring Point Source Discharge
While useful, surface water monitoring does not give insight into the source of the resistance genes, resistant pathogens, or medicine residues in the environment. Such insight could come from analyzing the places contaminants enter water, broadly classified as either point sources, which are single, identifiable entry points such as pharmaceutical factories or sewage treatment, and nonpoint sources, meaning coming from many sources diffusing through seepage or natural water cycling (EPA, 2021c; NOAA, 2021). The Clean Water Act requires monitoring at point sources, but discharge from nonpoint sources, including farms and feedlots, is not regulated (EPA, 2020b).
Given the uncertainty in measuring meaningful resistance traits in the environment, the way resistance-encoding genes move through the environment, and the sheer geographical scale involved, efforts to monitor resistance in the environment will likely have to start with a relatively narrow scope. Point source discharge is a good starting point given EPA’s statutory mandate to monitor them and their known association with resistance indicators (Fouz et al., 2020).
Wastewater treatment plants are a good entry point to monitor for resistance genes, resistant bacteria, and antimicrobial residues. This is pri-
marily because wastewater treatment is equipped to contain and remove water contaminants, but not to remove resistance genes or drug residues (Berendonk et al., 2015). Treatment plants typically discharge directly to aquatic environments, making them an important bridge between human-made contamination and the natural environment (Berendonk et al., 2015). Multiple studies have reported elevated levels of antimicrobial resistance downstream of wastewater discharges (Ashbolt et al., 2019; Zhang et al., 2009). Wastewater discharges also include waste streams from hospitals, which are of particular concern because of the antimicrobials residues and resistant pathogens found there. Research in Europe has shown indicators of antimicrobial resistance in wastewater largely parallel the relative burden of clinical resistance (Parnanen et al., 2019). Whether hospital effluent contributes disproportionately to the presence of antimicrobial resistance genes in wastewater treatment plants is as yet an unresolved scientific question, however (Buelow et al., 2018; Kraupner et al., 2021; Muller et al., 2018).
The monitoring of risk factors for resistance in manure would be particularly challenging. Given the broad variability when the manure is applied to fields and its runoff enters water, land application is typically classified as a nonpoint source. While certain kinds of animal agriculture such as contained animal feeding operations are regulated as point sources, monitoring waste collection facilities would likely be met with understandable pushback from farmers (EPA, 2020a). In most of the country there are too few farms even at the county level to make anonymization possible. Monitoring wastewater has the advantage of anonymity, and there is ample tie-in to resistance risk in agriculture as biosolids from treatment plants can be used to fertilize farm lands, bringing the readily exchangeable genetic information from resistant pathogens into contact with terrestrial organisms (Buta et al., 2021; EPA, 2021b; Mackie et al., 2006).
EPA had begun a project to validate hospital discharge as a high-strength source of antimicrobial resistance, looking specifically at ESBL E. coli and vancomycin-resistant Enterococcus spp. isolates and resistance genes, but this work was suspended because of the COVID-19 pandemic (Garland, 2020). The agency is now working with the CDC and the Department of Health and Human Services on surveillance for SARS-CoV-2 in wastewater, with plans to expand this system to look for resistant pathogens in the future (Anthes, 2021). Thus, as Box 4-3 described, the foundation on which to build surveillance is already in place.
Recommendation 4-2: The Environmental Protection Agency should provide guidance and funding to states for testing point source discharge at wastewater treatment plants for antimi
crobial resistance traits and integrating these data with other surveillance networks.
Monitoring wastewater discharge will provide a broader dataset to relate local environmental and clinical patterns of resistance. This includes relating indicators of antimicrobial resistance in waste to clinical resistance profiles and relating waste discharge to downstream resistance detected through other monitoring networks, such as EPA’s monitoring of streams and rivers. This monitoring will contribute to better linking of resistance indicators in waste to those found downstream in surface waters. Information from the proposed surveillance would, if included in the NCBI Pathogen Detection Database and the proposed NLM database described in Recommendation 4-1, contribute to a more complete mapping of the geographic distribution of resistance genes.
A recent EPA publication concluded that no one of the vast array of molecular methods available can fully characterize the burden of antimicrobial resistance in surface water (Franklin et al., 2021). A combination of genotypic and phenotypic tools will be needed in surveillance, at least until there is better consensus on what indicators best measure risk to human or animal health (Franklin et al., 2021). Bacterial culture may be best suited to link surface water resistance to animal or human infections, while molecular-genotypic approaches provide better insight into the movement of resistance genes and the transfer of resistance genes or traits to other bacteria (Franklin et al., 2021).
As a starting point, testing could focus on the subset of resistance genes and mobile genetic elements currently monitored in EPA surface water programs (Garland et al., 2018). Similar monitoring in Europe provided a useful baseline for understanding the relative abundance of key resistance genes in wastewater treatment and downstream (Cacace et al., 2019). It also allowed for identification of the resistance gene best suited to tracking as a proxy for resistance in the environment (blaOXA58) and shed light on wastewater treatment practices that can reduce the burden of resistance genes in the water supply (Cacace et al., 2019).5 It may take time to identify the analytic techniques best suited to identify the resistance genes or genetic elements in water that pose the most serious risk to human or animal health. In the short term, a combination of culture-based methods paired with genome sequencing and metagenomic analysis may be necessary (Franklin et al., 2021).
Some evidence indicates that the amount of resistant pathogens or genes discharged through wastewater treatment is directly related to the
___________________
5 A gene associated with carbapenem-resistance in Acinetobacter spp, blaOXA58, was isolated in clinical Proteus mirablis in 2017 (Girlich et al., 2017).
composition of the effluent discharged into the environment (Ju et al., 2019). Characterizing the relative abundance of resistance traits at this point source would be a valuable first step to understanding the extent to which water can amplify and convey antimicrobial resistance in the environment. The proposed surveillance of point source discharge would also make a solid foundation upon which EPA could build future water monitoring efforts. As better clarity about the monitoring of antimicrobial residues and concentrations in water emerge from research, this system could be expanded to monitor other resistance indicators in other water sources.
Understanding Water as a Conduit of Resistance
Monitoring waste streams and advancing analytical methods will help characterize the role of water as a conduit of antimicrobial resistance. This characterization may inform selection of water quality indicators. EPA provides state and local governments with water quality criteria for aquatic life, recreational and drinking water (EPA, 2020c). These include measures of fecal contamination as indicated by E. coli and Enterococcus species (EPA, 2015). Recreational water sites are regularly monitored during high-use seasons to ensure the criteria are met. EPA could eventually develop similar water quality criteria with threshold limits for antimicrobial resistance risk. There is evidence that ingesting resistant E. coli while surfing or swimming leads to colonization with the resistant pathogen (Leonard et al., 2018). The information gleaned from water surveillance could eventually feed in to the longer-term goal of monitoring recreational water for resistant pathogens or other indicators of resistance that pose risk to human health (Huijbers et al., 2019).
In the same way, it is possible that with sufficient evidence of health risk, EPA may eventually identify certain pathogens, medicines or residues, or resistance genes that qualify as pollutants that would need to be removed above a certain threshold. Removing antimicrobials at wastewater treatment plants is possible, though the efficiency of the process depends on both the physical and chemical properties of the medicine and the operating conditions at the treatment plant (Michael et al., 2013). Some evidence suggests that macrolide antimicrobials are difficult to remove from wastewater, though this may depend on the treatment methods used (Barbosa et al., 2021; Pan and Yau, 2021). There is evidence that current action at these plants can reduce the abundance of some resistance genes, though genes associated with resistance to vancomycin are notably unaffected (Parnanen et al., 2019). Older research indicated that resistance genes survive tertiary treatment and contribute to the burden of resistance genes in the environment (LaPara et al., 2011).
Lessons learned from mitigating resistance at point source discharge could then be applied to other discharge sites. Discharge from nonpoint sources would be monitored and mitigation strategies implemented through Section 319 of the Clean Water Act, which provides federal funding for state, tribal, and territorial authorities in their work to manage water quality at nonpoint sources (EPA, 2021a). Understanding the environmental fate and transport of antimicrobials and their metabolites, resistant organisms, and resistance genes, as well as their decay patterns, will be a precursor to any nonpoint source action against antimicrobial resistance. In particular, understanding environmental transport mechanisms will inform more efficient design of environmental monitoring systems.
REFERENCES
Aarestrup, F. M., and M. E. J. Woolhouse. 2020. Using sewage for surveillance of antimicrobial resistance. Science 367(6478):630-632.
Africa CDC (Centres for Disease Control and Prevention). 2020. US $100 million Africa pathogen genomics initiative to boost disease surveillance and emergency response capacity in Africa. Addis Ababa, Ethiopia: Africa Centres for Disease Control and Prevention.
AHRQ (Agency for Healthcare Research and Quality). 2014. Concise antibiogram toolkit using WHONET to create your antibiogram. https://www.ahrq.gov/sites/default/files/wysiwyg/professionals/quality-patient-safety/patient-safety-resources/resources/nh-aspguide/module2/toolkit1/cat_whonet.pdf (accessed July 30, 2021).
Alcock, B. P., A. R. Raphenya, T. T. Y. Lau, K. K. Tsang, M. Bouchard, A. Edalatmand, W. Huynh, A. V. Nguyen, A. A. Cheng, S. Liu, S. Y. Min, A. Miroshnichenko, H. K. Tran, R. E. Werfalli, J. A. Nasir, M. Oloni, D. J. Speicher, A. Florescu, B. Singh, M. Faltyn, A. Hernandez-Koutoucheva, A. N. Sharma, E. Bordeleau, A. C. Pawlowski, H. L. Zubyk, D. Dooley, E. Griffiths, F. Maguire, G. L. Winsor, R. G. Beiko, F. S. L. Brinkman, W. W. L. Hsaio, G. V. Domselaar, and A. G. McArthur. 2020. Card 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Research 48(D1):D517-D525.
Allen, H. K., J. Donato, H. H. Wang, K. A. Cloud-Hansen, J. Davies, and J. Handelsman. 2010. Call of the wild: Antibiotic resistance genes in natural environments. Nature Reviews: Microbiology 8(4):251-259.
AMR Industry Alliance. 2017a. Merck & Co., Inc.—study for monitoring antimicrobial resistance trends (SMART). https://www.amrindustryalliance.org/case-study/study-for-monitoring-antimicrobial-resistance-trends-smart (accessed July 30, 2021).
AMR Industry Alliance. 2017b. GSK—survey of antibiotic resistance (SOAR). https://www.amrindustryalliance.org/case-study/gsks-survey-of-antibiotic-resistance-soar (accessed July 30, 2021).
AMR Research Initiative. 2021a. Project overview. https://amr.theodi.org/project-overview (accessed July 30, 2021).
AMR Research Initiative. 2021b. About. https://amr.theodi.org (accessed July 27, 2021).
Anthes, E. 2021. From the wastewater drain, solid pandemic data. The New York Times, May 7 2021.
APHIS (Animal and Plant Health Inspection Service). 2021. APHIS’s American Rescue Plan (ARP) surveillance program: Strategic framework. https://www.aphis.usda.gov/publications/aphis_general/arp-strategic-framework.pdf (accessed September 17, 2021).
Ardal, C., D. McAdams, A. L. Wester, and S. Mogedal. 2021. Adapting environmental surveillance for polio to the need to track antimicrobial resistance. Bulletin of the World Health Organization 99(3):239-240.
Argimon, S., M. A. L. Masim, J. M. Gayeta, M. L. Lagrada, P. K. V. Macaranas, V. Cohen, M. T. Limas, H. O. Espiritu, J. C. Palarca, J. Chilam, M. C. Jamoralin, Jr., A. S. Villamin, J. B. Borlasa, A. M. Olorosa, L. F. T. Hernandez, K. D. Boehme, B. Jeffrey, K. Abudahab, C. M. Hufano, S. B. Sia, J. Stelling, M. T. G. Holden, D. M. Aanensen, and C. C. Carlos. 2020. Integrating whole-genome sequencing within the national antimicrobial resistance surveillance program in the Philippines. Nature Communications 11(1):2719.
Ashbolt, N., A. Pruden, J. Miller, M. V. Riquelme, A. Maile-Moskowitz, A. Pruden, N. Ashbolt, and J. Miller. 2019. Antimicrobal resistance: Fecal sanitation strategies for combating a global public health threat. In Water and sanitation for the 21st century: Health and microbiological aspects of excreta and wastewater management (Global Water Pathogens Project), edited by J. B. Rose and B. Jiménez-Cisneros. East Lansing, MI: Michigan State University.
Ashley, E. A., J. Recht, A. Chua, D. Dance, M. Dhorda, N. V. Thomas, N. Ranganathan, P. Turner, P. J. Guerin, N. J. White, and N. P. Day. 2018. An inventory of supranational antimicrobial resistance surveillance networks involving low- and middle-income countries since 2000. Journal of Antimicrobial Chemotherapy 73(7):1737-1749.
Baquero, F. 2001. Low-level antibacterial resistance: A gateway to clinical resistance. Drug Resistance Updates 4(2):93-105.
Barbosa, V., M. Morais, A. Silva, C. Delerue-Matos, S. A. Figueiredo, and V. F. Domingues. 2021. Comparison of antibiotic resistance in the influent and effluent of two wastewater treatment plants. AIMS Environmental Science 8(2):101-116.
Bard, J. D., and F. Lee. 2018. Why can’t we just use PCR? The role of genotypic versus phenotypic testing for antimicrobial resistance testing. Clinical Microbiology Newsletter 40(11):87-95.
Bengtsson-Palme, J., and D. G. Larsson. 2016. Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International 86:140-149.
Berendonk, T. U., C. M. Manaia, C. Merlin, D. Fatta-Kassinos, E. Cytryn, F. Walsh, H. Burgmann, H. Sorum, M. Norstrom, M. N. Pons, N. Kreuzinger, P. Huovinen, S. Stefani, T. Schwartz, V. Kisand, F. Baquero, and J. L. Martinez. 2015. Tackling antibiotic resistance: The environmental framework. Nature Reviews: Microbiology 13(5):310-317.
Biomerieux. 2020. Global point prevalence survey. https://amr.biomerieux.com/en/ourcommitment/global-point-prevalence-survey (accessed July 27, 2021).
Bortolaia, V., R. S. Kaas, E. Ruppe, M. C. Roberts, S. Schwarz, V. Cattoir, A. Philippon, R. L. Allesoe, A. R. Rebelo, A. F. Florensa, L. Fagelhauer, T. Chakraborty, B. Neumann, G. Werner, J. K. Bender, K. Stingl, M. Nguyen, J. Coppens, B. B. Xavier, S. Malhotra-Kumar, H. Westh, M. Pinholt, M. F. Anjum, N. A. Duggett, I. Kempf, S. Nykasenoja, S. Olkkola, K. Wieczorek, A. Amaro, L. Clemente, J. Mossong, S. Losch, C. Ragimbeau, O. Lund, and F. M. Aarestrup. 2020. Resfinder 4.0 for predictions of phenotypes from genotypes. Journal of Antimicrobial Chemotherapy 75(12):3491-3500.
Brinch, C., and F. M. Aarestrup. 2020. Global sewage situation report. The Global Sewage Surveillance Project Consortium. https://www.compare-europe.eu/-/media/Sites/Global_Surveillance/Global-Sewage-Updates/Global-sewage-situation-report-May-2020.ashx?la=da&hash=16E13867C2D71C3AE08E71E53CFE8F891D5F2ABE (accessed September 24, 2021).
Buelow, E., J. R. Bayjanov, E. Majoor, R. J. Willems, M. J. Bonten, H. Schmitt, and W. van Schaik. 2018. Limited influence of hospital wastewater on the microbiome and resistome of wastewater in a community sewerage system. FEMS Microbiology Ecology 94(7).
Buelow, E., A. Rico, M. Gaschet, J. Lourenco, S. P. Kennedy, L. Wiest, M. C. Ploy, and C. Dagot. 2020. Hospital discharges in urban sanitation systems: Long-term monitoring of wastewater resistome and microbiota in relationship to their eco-exposome. Water Research X 7:100045.
Buta, M., J. Hubeny, W. Zielinski, M. Harnisz, and E. Korzeniewska. 2021. Sewage sludge in agriculture: The effects of selected chemical pollutants and emerging genetic resistance determinants on the quality of soil and crops—a review. Ecotoxicology Environmental Safety 214:112070.
Cacace, D., D. Fatta-Kassinos, C. M. Manaia, E. Cytryn, N. Kreuzinger, L. Rizzo, P. Karaolia, T. Schwartz, J. Alexander, C. Merlin, H. Garelick, H. Schmitt, D. de Vries, C. U. Schwermer, S. Meric, C. B. Ozkal, M. N. Pons, D. Kneis, and T. U. Berendonk. 2019. Antibiotic resistance genes in treated wastewater and in the receiving water bodies: A pan-European survey of urban settings. Water Research 162:320-330.
Cappuccino, J. G., and N. Sherman. 2014. Microbiology: A laboratory manual, 10th ed. New York: Pearson Education.
CDC (Centers for Disease Control and Prevention). 2019a. Core elements of hospital antibiotic stewardship programs. Atlanta, GA: Centers for Disease Control and Prevention.
CDC. 2019b. Laboratory testing. https://www.cdc.gov/mrsa/lab/index.html#anchor_1548442775 (accessed July 30, 2021).
CDC. 2021a. Gonococcal isolate surveillance project (GISP). https://www.cdc.gov/std/gisp/default.htm (accessed September 17, 2021).
CDC. 2021b. Waterborne disease & outbreak surveillance reporting: Wastewater surveillance. https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/wastewater-surveillance.html (accessed June 30, 2021).
CDDEP (Center for Disease Dynamics, Economics & Policy). 2021. Resistancemap: Antibiotic resistance. https://resistancemap.cddep.org/AntibioticResistance.php (accessed September 14, 2021).
Chandrasekera, R. M., E. P. Lesho, U. Chukwuma, J. F. Cummings, and P. E. Waterman. 2015. The state of antimicrobial resistance surveillance in the military health system: A review of improvements made in the last 10 years and remaining surveillance gaps. Military Medicine 180(2):145-150.
CLSI (Clinical and Laboratory Standards Institute). 2014. Analysis and presentation of cumulative antimicrobial susceptibility test data; approved guideline, 4th ed. Wayne, PA: Clinical Laboratory Standards Institute. https://www.nih.org.pk/wp-content/uploads/2021/02/CLSI-M39A4E-Antibiogram.pdf (accessed September 22, 2021).
Cusack, T. P., E. A. Ashley, C. L. Ling, S. Rattanavong, T. Roberts, P. Turner, T. Wangrangsimakul, and D. A. B. Dance. 2019. Impact of CLSI and EUCAST breakpoint discrepancies on reporting of antimicrobial susceptibility and AMR surveillance. Clinical Microbiology and Infection 25(7):910-911.
Dantas, G., M. O. Sommer, R. D. Oluwasegun, and G. M. Church. 2008. Bacteria subsisting on antibiotics. Science 320(5872):100-103.
Davis, G. S., K. Waits, L. Nordstrom, B. Weaver, M. Aziz, L. Gauld, H. Grande, R. Bigler, J. Horwinski, S. Porter, M. Stegger, J. R. Johnson, C. M. Liu, and L. B. Price. 2015. Intermingled klebsiella pneumoniae populations between retail meats and human urinary tract infections. Clinical Infectious Diseases 61(6):892-899.
Diallo, O. O., S. A. Baron, C. Abat, P. Colson, H. Chaudet, and J. M. Rolain. 2020. Antibiotic resistance surveillance systems: A review. Journal of Global Antimicrobial Resistance 23:430-438.
Dien Bard, J., and E. McElvania. 2020. Panels and syndromic testing in clinical microbiology. Clinics in Laboratory Medicine 40(4):393-420.
Dunne, W. M., Jr., M. Jaillard, O. Rochas, and A. Van Belkum. 2017. Microbial genomics and antimicrobial susceptibility testing. Expert Review of Molecular Diagnostics 17(3):257-269.
Enne, V. I., A. P. Johnson, and J. D. Perry. 2016. Preface. Journal of Antimicrobial Chemotherapy 71(Suppl 1):i1.
EPA (Environmental Protection Agency). 2015. Recreational water quality criteria. Environmental Protection Agency. https://www.epa.gov/sites/default/files/2015-10/documents/rwqc2012.pdf (accessed July 30, 2021).
EPA. 2020a. Animal feeding operations (AFOS). https://www.epa.gov/npdes/animal-feeding-operations-afos (accessed July 30, 2021).
EPA. 2020b. Summary of the Clean Water Act. https://www.epa.gov/laws-regulations/summary-clean-water-act (accessed July 20, 2021).
EPA. 2020c. Water quality criteria. https://www.epa.gov/wqc (accessed July 30, 2021).
EPA. 2021a. 319 grant: Current guidance. https://www.epa.gov/nps/319-grant-current-guidance (accessed October 11, 2021).
EPA. 2021b. Basic information about biosolids. https://www.epa.gov/biosolids/basic-information-about-biosolids (accessed July 30, 2021).
EPA. 2021c. Basic information about nonpoint source (NPS) pollution. https://www.epa.gov/nps/basic-information-about-nonpoint-source-nps-pollution (accessed July 30, 2021).
FAO (Food and Agriculture Organization of the United Nations). 2019a. Monitoring and surveillance of antimicrobial resistance in bacteria from healthy food animals intended for consumption. In Regional Antimicrobial Resistance Monitoring and Surveillance Guidelines. Bangkok, Thailand: Food and Agriculture Organization of the United Nations.
FAO. 2019b. Monitoring and surveillance of antimicrobial resistance in bacteria from healthy food animals intended for consumption. In Regional Antimicrobial Resistance Monitoring and Surveillance Guidelines—Volume 1. Bangkok, Thailand: Food and Agriculture Organization of the United Nations.
FAO. 2021a. Evaluation of FAO’s role and work on antimicrobial resistance (AMR). In Thematic Evaluation Series. Rome, Italy: Food and Agriculture Organization of the United Nations.
FAO. 2021b. FAO action plan on antimicrobial resistance 2021-2025. Food and Agriculture Organization of the United Nations. https://www.fao.org/3/ne859en/ne859en.pdf (accessed July 30, 2021).
FAO. 2021c. FAO and WHO activities to support monitoring and surveillance of antimicrobial resistance in the food and agriculture sectors. http://www.fao.org/fao-who-codexalimentarius/news-and-events/news-details/en/c/1381093 (accessed July 30, 2021).
FAO and UN (United Nations). 2021. Antimicrobial resistance. http://www.fao.org/antimicrobial-resistance/en (accessed September 22, 2021).
FAO and WHO. 2019. Joint FAO/WHO food standards programme—ad hoc codex intergovernmental task force on antimicrobial resistance. http://www.fao.org/fao-who-codexalimentarius/sh-proxy/en/?lnk=1&url=https%253A%252F%252Fworkspace.fao.org%252Fsites%252Fcodex%252FMeetings%252FCX-804-07%252FWDs%252Famr07_03e.pdf (accessed July 30, 2021).
FAO and WHO. 2021. Request for comments on the guidelines on integrated monitoring and surveillance of foodborne antimicrobial resistance (at step 3). http://www.fao.org/fao-who-codexalimentarius/sh-proxy/en/?lnk=1&url=https%253A%252F%252Fworkspace.fao.org%252Fsites%252Fcodex%252FCircular%252520Letters%252FCL%2525202021-33-OCS%252Fcl21_33e.pdf (accessed July 30, 2021).
FAO, OIE (World Organisation for Animal Health), and WHO (World Health Organization). 2020. Annual progress report 2020- antimicrobial resistance multi-partner trust fund. Geneva, Switzerland: World Health Organization.
FDA (Food and Drug Administration). 2021. 2020 virtual public meeting of the national antimicrobial resistance monitoring system. https://www.regulations.gov/document/FDA-2020-N-1764-0017#collapseAttachmentMetadata-ember21 (accessed July 30, 2021).
Feldgarden, M., V. Brover, N. Gonzalez-Escalona, J. G. Frye, J. Haendiges, D. H. Haft, M. Hoffmann, J. B. Pettengill, A. B. Prasad, G. E. Tillman, G. H. Tyson, and W. Klimke. 2021a. Amrfinderplus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Scientific Reports 11(1):12728.
Feldgarden, M., V. B., B. Fedorov, D. H. Haft, A. B. Prasad, W. Klimke. 2021b. NCBI’s pathogen detection system and MICROBIGG-E enable evolutionary analysis of aac(6’)-ib family aminoglycoside modifying enzymes. World Microbe Forum. https://www.abstractsonline.com/pp8/#!/9286/presentation/8546 (accessed June 30, 2021).
Fouz, N., K. N. A. Pangesti, M. Yasir, A. L. Al-Malki, E. I. Azhar, G. A. Hill-Cawthorne, and M. Abd El Ghany. 2020. The contribution of wastewater to the transmission of antimicrobial resistance in the environment: Implications of mass gathering settings. Tropical Medicine and Infectious Disease 5(1):33.
Franklin, A. M., N. E. Brinkman, M. A. Jahne, and S. P. Keely. 2021. Twenty-first century molecular methods for analyzing antimicrobial resistance in surface waters to support one health assessments. Journal of Microbiological Methods 184:106174.
Franzosa, E. A., X. C. Morgan, N. Segata, L. Waldron, J. Reyes, A. M. Earl, G. Giannoukos, M. R. Boylan, D. Ciulla, D. Gevers, J. Izard, W. S. Garrett, A. T. Chan, and C. Huttenhower. 2014. Relating the metatranscriptome and metagenome of the human gut. Proceedings of the National Academy of Sciences of the United States of America 111(22):E2329-E2338.
Fridkin, S. K., A. A. Cleveland, I. See, and R. Lynfield. 2015. Emerging infections program as surveillance for antimicrobial drug resistance. Emerging Infectious Diseases 21(9):1578-1581.
Fuhrmeister, A. S., and R. N. Jones. 2019. The importance of antimicrobial resistance monitoring worldwide and the origins of SENTRY antimicrobial surveillance program. Open Forum Infectious Diseases 6(Suppl 1):S1-S4.
Galhano, B. S. P., R. G. Ferrari, P. Panzenhagen, A. C. S. de Jesus, and C. A. Conte-Junior. 2021. Antimicrobial resistance gene detection methods for bacteria in animal-based foods: A brief review of highlights and advantages. Microorganisms 9(5):923.
GAO (Government Accountability Office). 2020. Antibiotic resistance: Additional federal actions needed to better determine magnitude and reduce impact. Washington, DC: Government Accountability Office.
GAO. 2021. Science & tech spotlight: Genomic sequencing of infectious pathogens. https://www.gao.gov/products/gao-21-426sp (accessed September 17, 2021).
Garland, J. 2020. NARMS surface water pilot overview. Annual Meeting of the National Institute of Animal Agriculture, Cincinnati, Ohio. https://cfpub.epa.gov/si/si_public_record_Report.cfm?dirEntryId=350198&Lab=CESER (accessed July 30, 2021).
Garland, J. L., N. Brinkman, S. Keely, and M. Jahne. 2018. Geospatial distribution of antimicrobial resistance genes in us rivers and streams. https://www.hhs.gov/sites/default/files/garland-paccarb.pdf (accessed July 30, 2021).
Girlich, D., R. A. Bonnin, P. Bogaerts, M. De Laveleye, D. T. Huang, L. Dortet, P. Glaser, Y. Glupczynski, and T. Naas. 2017. Chromosomal amplification of the blaoxa-58 carbapenemase gene in a proteus mirabilis clinical isolate. Antimicrobial Agents and Chemotherapy 61(2):e01697-01616.
Global PPS. 2021a. About the global PPS project. https://www.global-pps.com/project (accessed July 30, 2021).
Global PPS. 2021b. Acknowledgements. https://www.global-pps.com/acknowledgements (accessed July 27, 2021).
Harris, S. R., E. J. Cartwright, M. E. Torok, M. T. Holden, N. M. Brown, A. L. Ogilvy-Stuart, M. J. Ellington, M. A. Quail, S. D. Bentley, J. Parkhill, and S. J. Peacock. 2013. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant staphylococcus aureus: A descriptive study. Lancet Infectious Diseases 13(2):130-136.
Harris-Lovett, S., K. Nelson, P. Beamer, H. N. Bischel, A. Bivins, A. Bruder, C. Butler, T. D. Camenisch, S. K. De Long, S. Karthikeyan, D. A. Larsen, K. Meierdiercks, P. Mouser, S. Pagsuyoin, S. Prasek, T. S. Radniecki, J. L. Ram, D. K. Roper, H. Safford, S. P. Sherchan, W. Shuster, T. Stalder, R. T. Wheeler, and K. S. Korfmacher. 2021. Wastewater surveillance for SAR-CoV-2 on college campuses: Initial efforts, lessons learned and research needs. medRxiv 18(9).
Health.mil. 2021. Global emerging infections surveillance. health.mil/Military-Health-Topics/Combat-Support/Armed-Forces-Health-Surveillance-Branch/Global-Emerging-Infections-Surveillance-and-Response (accessed September 17, 2021).
Hendriksen, R. S., V. Bortolaia, H. Tate, G. H. Tyson, F. M. Aarestrup, and P. F. McDermott. 2019a. Using genomics to track global antimicrobial resistance. Front Public Health 7(242):242.
Hendriksen, R. S., P. Munk, P. Njage, B. van Bunnik, L. McNally, O. Lukjancenko, T. Roder, D. Nieuwenhuijse, S. K. Pedersen, J. Kjeldgaard, R. S. Kaas, P. Clausen, J. K. Vogt, P. Leekitcharoenphon, M. G. M. van de Schans, T. Zuidema, A. M. de Roda Husman, S. Rasmussen, B. Petersen, The Global Sewage Surveillance Project Consortium, C. Amid, G. Cochrane, T. Sicheritz-Ponten, H. Schmitt, J. R. M. Alvarez, A. Aidara-Kane, S. J. Pamp, O. Lund, T. Hald, M. Woolhouse, M. P. Koopmans, H. Vigre, T. N. Petersen, and R. M. Aarestrup. 2019b. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nature Communications 10(1):1124.
Hermsen, E. D., R. L. Sibbel, and S. Holland. 2020. The role of pharmaceutical companies in antimicrobial stewardship: A case study. Clinical Infectious Diseases 71(3):677-681.
Huijbers, P. M. C., C. F. Flach, and D. G. J. Larsson. 2019. A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environment International 130:104880.
INICC (International Nosocomial Infection Control Consortium). 2013. Multicentric network of cities and countries. http://www.inicc.org/multicentric (accessed July 27, 2021).
Iskandar, K., L. Molinier, S. Hallit, M. Sartelli, T. C. Hardcastle, M. Haque, H. Lugova, S. Dhingra, P. Sharma, S. Islam, I. Mohammed, I. Naina Mohamed, P. A. Hanna, S. E. Hajj, N. A. H. Jamaluddin, P. Salameh, and C. Roques. 2021. Surveillance of antimicrobial resistance in low- and middle-income countries: A scattered picture. Antimicrobial Resistance & Infection Control 10(1):63.
Ju, F., K. Beck, X. Yin, A. Maccagnan, C. S. McArdell, H. P. Singer, D. R. Johnson, T. Zhang, and H. Burgmann. 2019. Wastewater treatment plant resistomes are shaped by bacterial composition, genetic exchange, and upregulated expression in the effluent microbiomes. ISME Journal 13(2):346-360.
Keshaviah, A., X. C. Hu, and M. Henry. 2021. Developing a flexible national wastewater surveillance system for COVID-19 and beyond. Environmental Health Perspectives 129(4):45002.
Korry, B. J., D. J. Cabral, and P. Belenky. 2020. Metatranscriptomics reveals antibiotic-induced resistance gene expression in the murine gut microbiota. Frontiers in Microbiology 11:322.
Koser, C. U., M. J. Ellington, and S. J. Peacock. 2014. Whole-genome sequencing to control antimicrobial resistance. Trends in Genetics 30(9):401-407.
Kowalchuk, G. A., A. G. Speksnijder, K. Zhang, R. M. Goodman, and J. A. van Veen. 2007. Finding the needles in the metagenome haystack. Microbial Ecology 53(3):475-485.
Kraupner, N., M. Hutinel, K. Schumacher, D. A. Gray, M. Genheden, J. Fick, C. F. Flach, and D. G. J. Larsson. 2021. Evidence for selection of multi-resistant E. coli by hospital effluent. Environment International 150:106436.
Kreier, F. 2021. The myriad ways sewage surveillance is helping fight COVID around the world. Nature. https://www.nature.com/articles/d41586-021-01234-1 (accessed September 22, 2021).
LaPara, T. M., T. R. Burch, P. J. McNamara, D. T. Tan, M. Yan, and J. J. Eichmiller. 2011. Tertiary-treated municipal wastewater is a significant point source of antibiotic resistance genes into Duluth-Superior harbor. Environmental Science & Technology 45(22):9543-9549.
Lee, L. M., S. M. Teutsch, S. B. Thacker, and M. E. St. Louis. 2010. Principles & practice of public health surveillance. New York: Oxford University Press.
Leitmeyer, K. C., L. Espinosa, E. K. Broberg, M. J. Struelens, ECDC National Focal Points Laboratory E-reporting Survey Group Members, and ECDC National Focal Points Laboratory E-reporting Survey Group. 2020. Automated digital reporting of clinical laboratory information to national public health surveillance systems, results of a EU/EEA survey, 2018. Euro Surveillance 25(39):1900591.
Leonard, A. F. C., L. Zhang, A. J. Balfour, R. Garside, P. M. Hawkey, A. K. Murray, O. C. Ukoumunne, and W. H. Gaze. 2018. Exposure to and colonisation by antibiotic-resistant E. coli in UK coastal water users: Environmental surveillance, exposure assessment, and epidemiological study (beach bum survey). Environment International 114:326-333.
Luby, E., A. M. Ibekwe, J. Zilles, and A. Pruden. 2016. Molecular methods for assessment of antibiotic resistance in agricultural ecosystems: Prospects and challenges. Journal of Environmental Quality 45(2):441-453.
Lumen. 2021. Testing the effectiveness of antimicrobials. https://courses.lumenlearning.com/microbiology/chapter/testing-the-effectiveness-of-antimicrobials (accessed May 19, 2021).
Mackie, R. I., S. Koike, I. Krapac, J. Chee-Sanford, S. Maxwell, and R. I. Aminov. 2006. Tetracycline residues and tetracycline resistance genes in groundwater impacted by swine production facilities. Animal Biotechnology 17(2):157-176.
Maki, A., P. Salmi, A. Mikkonen, A. Kremp, and M. Tiirola. 2017. Sample preservation, DNA or RNA extraction and data analysis for high-throughput phytoplankton community sequencing. Frontiers in Microbiology 8:1848.
Manaia, C. M. 2017. Assessing the risk of antibiotic resistance transmission from the environment to humans: Non-direct proportionality between abundance and risk. Trends in Microbiology 25(3):173-181.
Martinez, J. L., T. M. Coque, and F. Baquero. 2015. What is a resistance gene? Ranking risk in resistomes. Nature Reviews: Microbiology 13(2):116-123.
McLain, J. E., E. Cytryn, L. M. Durso, and S. Young. 2016. Culture-based methods for detection of antibiotic resistance in agroecosystems: Advantages, challenges, and gaps in knowledge. Journal of Environmental Quality 45(2):432-440.
Meyer, W. G., J. A. Pavlin, D. Hospenthal, C. K. Murray, K. Jerke, A. Hawksworth, D. Metzgar, T. Myers, D. Walsh, M. Wu, R. Ergas, U. Chukwuma, S. Tobias, J. Klena, I. Nakhla, M. Talaat, R. Maves, M. Ellis, G. Wortmann, D. L. Blazes, and L. Lindler. 2011. Antimicrobial resistance surveillance in the AFHSC-GEIS network. BMC Public Health 11(Suppl 2):S8.
Michael, I., L. Rizzo, C. S. McArdell, C. M. Manaia, C. Merlin, T. Schwartz, C. Dagot, and D. Fatta-Kassinos. 2013. Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: A review. Water Research 47(3):957-995.
Minnesota Department of Health. 2015. About antibiograms—antimicrobial susceptibilities of selected pathogens. Minnesota Department of Health. https://www.health.state.mn.us/diseases/antibioticresistance/abx/antibiograms.pdf (accessed September 22, 2021).
Muller, H., E. Sib, M. Gajdiss, U. Klanke, F. Lenz-Plet, V. Barabasch, C. Albert, A. Schallenberg, C. Timm, N. Zacharias, R. M. Schmithausen, S. Engelhart, M. Exner, M. Parcina, C. Schreiber, and G. Bierbaum. 2018. Dissemination of multi-resistant gram-negative bacteria into German wastewater and surface waters. FEMS Microbiology Ecology 94(5).
Murray, A. K., I. Stanton, W. H. Gaze, and J. Snape. 2021. Dawning of a new era: Environmental risk assessment of antibiotics and their potential to select for antimicrobial resistance. Water Research 200:117233.
NCBI (National Center for Biotechnology Information). 2019. NCBI pathogen detection. https://ftp.ncbi.nlm.nih.gov/pub/factsheets/Factsheet_Pathogen_Detection.pdf (accessed July 30, 2021).
NIHR (National Institute for Health Research) Global Health Research Unit on Genomic Surveillance of AMR. 2020. Whole-genome sequencing as part of national and international surveillance programmes for antimicrobial resistance: A roadmap. BMJ Global Health 5(11):e002244.
NLM. n.d.a. (National Library of Medicine). About the NCBI pathogen detection system. https://www.ncbi.nlm.nih.gov/pathogens/about (accessed June 30, 2021).
NLM. n.d.b. Major contributors. https://www.ncbi.nlm.nih.gov/pathogens/contributors (accessed June 30, 2021).
NLM. n.d.c. National Database of Antibiotic Resistant Organisms (NDARO). https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance (accessed June 30, 2021).
NLM. 2018. Introducing the NCBI pathogen detection isolates browser. YouTube. https://www.youtube.com/watch?v=T8HluuzOb5c&t=133s (accessed September 22, 2021).
NLM. 2019a. AMRFinderPlus. https://www.ncbi.nlm.nih.gov/pathogens/antimicrobialresistance/AMRFinder (accessed June 30, 2021).
NLM. 2019b. Pathogen detection. https://www.ncbi.nlm.nih.gov/pathogens (accessed June 30, 2021).
NLM. 2020. Pathogen detection help document. https://www.ncbi.nlm.nih.gov/pathogens/pathogens_help/#amr-resources-schematic (accessed June 30, 2021).
NLM.2021a.Howtosubmitdataforreal-timeanalysis. https://www.ncbi.nlm.nih.gov/pathogens/submit-data (accessed June 30, 2021).
NLM. 2021b. Organism groups. https://www.ncbi.nlm.nih.gov/pathogens/organisms (accessed June 30, 2021).
NOAA (National Oceanic and Atmospheric Administration). 2021. Point source pollution tutorial. https://oceanservice.noaa.gov/education/tutorial_pollution/03pointsource.html (accessed July 30, 2021).
Norman, R. S. 2020. AMR and SARS-CoV-2 surveillance in sewage. 14th Public Virtual Meeting of the Presidential Advisory Council on Combating Antibiotic-Resistant Bacteria, September 10, 2020. https://www.hhs.gov/sites/default/files/norman-wastewater-surveillance.pdf (accessed July 30, 2021).
OPHSS (Office of Public Health Scientific Services) and CDC. 2018. Public health surveillance: Preparing for the future. Atlanta, GA: Centers for Disease Control and Prevention.
Pan, M., and P. C. Yau. 2021. Fate of macrolide antibiotics with different wastewater treatment technologies. Water, Air, and Soil Pollution 232(3):103.
Parnanen, K. M. M., C. Narciso-da-Rocha, D. Kneis, T. U. Berendonk, D. Cacace, T. T. Do, C. Elpers, D. Fatta-Kassinos, I. Henriques, T. Jaeger, A. Karkman, J. L. Martinez, S. G. Michael, I. Michael-Kordatou, K. O’Sullivan, S. Rodriguez-Mozaz, T. Schwartz, H. Sheng, H. Sorum, R. D. Stedtfeld, J. M. Tiedje, S. V. D. Giustina, F. Walsh, I. Vaz-Moreira, M. Virta, and C. M. Manaia. 2019. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Science Advances 5(3):eaau9124.
Pascucci, M., G. Royer, J. Adamek, M. A. Asmar, D. Aristizabal, L. Blanche, A. Bezzarga, G. Boniface-Chang, A. Brunner, C. Curel, G. Dulac-Arnold, R. M. Fakhri, N. Malou, C. Nordon, V. Runge, F. Samson, E. Sebastian, D. Soukieh, J. P. Vert, C. Ambroise, and M. A. Madoui. 2021. AI-based mobile application to fight antibiotic resistance. Nature Communications 12(1):1173.
Pfizer. 2021. Antimicrobial surveillance. https://www.pfizer.com/science/therapeutic-areas/anti-infectives/antimicrobial-surveillance (accessed August 5, 2021).
Polianciuc, S. I., A. E. Gurzau, B. Kiss, M. G. Stefan, and F. Loghin. 2020. Antibiotics in the environment: Causes and consequences. Medicine and Pharmacy Reports 93(3):231-240.
Prasad, A. B., V. B., M. Feldgarden, B. Fedorov, D. H. Haft, W. Klimke. 2021. NCBI’s AMRFinderPlus, isolates Browser, and MicroBIGG-E identify quinolone resistance mutations and their sources. World Microbe Forum. https://www.abstractsonline.com/pp8/#!/9286/presentation/3194 (accessed September 22, 2021).
Pruden, A., R. E. Alcalde, P. J. J. Alvarez, N. Ashbolt, H. Bischel, N. L. Capiro, E. Crossette, D. Frigon, K. Grimes, C. N. Haas, K. Ikuma, A. Kappell, T. LaPara, L. Kimbell, M. Li, X. Li, P. McNamara, Y. Seo, M. D. Sobsey, E. Sozzi, T. Navab-Daneshmand, L. Raskin, M. V. Riquelme, P. Vikesland, K. Wigginton, and Z. Zhou. 2018. An environmental science and engineering framework for combating antimicrobial resistance. Environmental Engineering Science 35(10):1005-1011.
Ranjalkar, J., and S. J. Chandy. 2019. India’s national action plan for antimicrobial resistance—an overview of the context, status, and way ahead. Journal of Family Medicine and Primary Care 8(6):1828-1834.
Rosenthal, V. D. 2016. International nosocomial infection control consortium (INICC) resources: INICC multidimensional approach and INICC surveillance online system. American Journal of Infection Control 44(6):e81-e90.
Rothrock, M. J., P. L. Keen, K. L. Cook, L. M. Durso, A. M. Franklin, and R. S. Dungan. 2016. How should we be determining background and baseline antibiotic resistance levels in agroecosystem research? Journal of Environmental Quality 45(2):420-431.
Ruzante, J. M., K. Olin, B. Munoz, J. Nawrocki, R. Selvarangan, and L. Meyers. 2021. Real-time gastrointestinal infection surveillance through a cloud-based network of clinical laboratories. PloS One 16(4):e0250767.
Saadat, S., K. Solhjoo, M. J. Norooz-Nejad, and A. Kazemi. 2014. VanA and vanB positive vancomycin-resistant staphylococcus aureus among clinical isolates in Shiraz, south of Iran. Oman Medical Journal 29(5):335-339.
Sayers, E. W., J. Beck, E. E. Bolton, D. Bourexis, J. R. Brister, K. Canese, D. C. Comeau, K. Funk, S. Kim, W. Klimke, A. Marchler-Bauer, M. Landrum, S. Lathrop, Z. Lu, T. L. Madden, N. O’Leary, L. Phan, S. H. Rangwala, V. A. Schneider, Y. Skripchenko, J. Wang, J. Ye, B. W. Trawick, K. D. Pruitt, and S. T. Sherry. 2021. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research 49(D1):D10-D17.
Schmieder, R., and R. Edwards. 2012. Insights into antibiotic resistance through metagenomic approaches. Future Microbiology 7(1):73-89.
Schurch, A. C., and W. van Schaik. 2017. Challenges and opportunities for whole-genome sequencing-based surveillance of antibiotic resistance. Annals of the New York Academy of Sciences 1388(1):108-120.
Seale, A. C., N. C. Gordon, J. Islam, S. J. Peacock, and J. A. G. Scott. 2017. AMR surveillance in low and middle-income settings—a roadmap for participation in the global antimicrobial surveillance system (glass). Wellcome Open Research 2:92.
Shakya, M., C. C. Lo, and P. S. G. Chain. 2019. Advances and challenges in metatranscriptomic analysis. Frontiers in Genetics 10:904.
Strain, E. 2021. Machine learning for MIC predictions using whole-genome sequencing for FDA NARMS. World Microbe Forum. https://www.abstractsonline.com/pp8/#!/9286/presentation/2568 (accessed June 30, 2021).
Sundsfjord, A., G. S. Simonsen, B. C. Haldorsen, H. Haaheim, S. O. Hjelmevoll, P. Littauer, and K. H. Dahl. 2004. Genetic methods for detection of antimicrobial resistance. APMIS 112(11-12):815-837.
The White House. 2015. National action plan for combating antibiotic resistant bacteria. Washington, DC: The White House.
Tiwari, H. K., A. K. Das, D. Sapkota, K. Sivrajan, and V. K. Pahwa. 2009. Methicillin resistant staphylococcus aureus: Prevalence and antibiogram in a tertiary care hospital in Western Nepal. Journal of Infection in Developing Countries 3(9):681-684.
Torumkuney, D., R. Chaiwarith, W. Reechaipichitkul, K. Malatham, V. Chareonphaibul, C. Rodrigues, D. S. Chitins, M. Dias, S. Anandan, S. Kanakapura, Y. J. Park, K. Lee, H. Lee, J. Y. Kim, Y. Lee, H. K. Lee, J. H. Kim, T. Y. Tan, Y. X. Heng, P. Mukherjee, and I. Morrissey. 2016. Results from the Survey of Antibiotic Resistance (SOAR) 2012-14 in Thailand, India, South Korea and Singapore. Journal of Antimicrobial Chemotherapy 71(Suppl 1):i3-i19.
UN MPTF (United Nations Multi-Partner Trust Fund) Office. 2020. 2020 annual report UN multi-partner trust fund office. United Nations Center of Expertise on Pooled Financing Instruments for the Sustainable Development Goals. https://mptf.undp.org/document/download/26724 (accessed July 28, 2021).
Vaz-Moreira, I., O. C. Nunes, and C. M. Manaia. 2014. Bacterial diversity and antibiotic resistance in water habitats: Searching the links with the human microbiome. FEMS Microbiology Reviews 38(4):761-778.
Vikesland, P. J., A. Pruden, P. J. J. Alvarez, D. Aga, H. Burgmann, X. D. Li, C. M. Manaia, I. Nambi, K. Wigginton, T. Zhang, and Y. G. Zhu. 2017. Toward a comprehensive strategy to mitigate dissemination of environmental sources of antibiotic resistance. Environmental Science & Technology 51(22):13061-13069.
Vivli Center for Global Clinical Research Data. 2021. News & events. https://vivli.org/vivli-awarded-contract-to-improve-access-to-antimicrobial-resistance-surveillance-data (accessed July 27, 2021).
Vong, S., A. Anciaux, A. Hulth, J. Stelling, V. Thamlikitkul, S. Gupta, J. M. Fuks, K. Walia, P. Rattanumpawan, S. Eremin, K. Tisocki, T. R. Sedai, and A. Sharma. 2017. Using information technology to improve surveillance of antimicrobial resistance in Southeast Asia. BMJ 358:j3781.
Welcomme Trust. 2018. Initiatives for addressing antimicrobial resistance in the environment—current situation and challenges. https://wellcome.ac.uk/sites/default/files/antimicrobial-resistance-environment-report.pdf (accessed April 28, 2021).
WHO (World Health Organization). 2014. Antimicrobial resistance—global report on surveillance. Geneva, Switzerland: World Health Organization. https://apps.who.int/iris/bitstream/handle/10665/112642/9789241564748_eng.pdf?sequence=1 (accessed June 30, 2021).
WHO. 2015. Global antimicrobial resistance surveillance system—manual for early implementation. Geneva, Switzerland: World Health Organization.
WHO. 2017. Global antimicrobial resistance surveillance system (GLASS) report—early implementation. Geneva, Switzerland: World Health Organization
WHO. 2021a. Global antimicrobial resistance and use surveillance system (GLASS) report: 2021. Geneva, Switzerland: World Health Organization.
WHO. 2021b. Tripartite integrated system for surveillance on AMR and antimicrobial use (TISSA). https://www.ungm.org/Public/Notice/124127 (accessed July 30, 2021).
WHO. 2021c. Who integrated global surveillance on ESBL-producing E. coli using a “One Health” approach. Geneva, Switzerland: World Health Organization.
WHONET. 2020. WHONET for caesar: Manual. https://www.whonet.org/docserv/Manual_WHONET_for_CAESAR.pdf (accessed July 28, 2021).
WHONET. 2021. The microbiology laboratory database software. https://whonet.software.informer.com (accessed July 30, 2021).
Wright, G. D. 2010. The antibiotic resistome. Expert Opinion on Drug Discovery 5(8):779-788.
Zhang, X. X., T. Zhang, and H. H. Fang. 2009. Antibiotic resistance genes in water environment. Applied Microbiology and Biotechnology 82(3):397-414.
















































