10
Molecular Genetics of Speciation and Human Origins
Francisco J. Ayala, Anaías Escalante, Colm O'Huigin, and Jan Klein
Species are populations of organisms reproductively isolated from other organisms. Speciation is the process by which two gene pools, say X and Y, derived from an ancestral pool, say Z, acquire species-specific genes that keep them from interbreeding. The ancestral species may persist, so that speciation may simply involve the divergence of a new species, say X, from the ancestral one, Z.
Species continuously evolve; after a time, a descendant gene pool, say Z', may be sufficiently different from the ancestral gene pool, Z, as to be considered a different species. Whether or not Z and Z' would exhibit reproductive isolation from each other, and thus meet the criterion for speciation, is an empirically meaningless question since the matter cannot be tested. But ancestral and descendant populations are given different species names if they are as different from each other as other contemporary species—for example, if Z' is as different from Z as it is from X or Y.
The process of speciation is notoriously refractory to investigation because it is complex and usually takes a long time, and also because speciation can happen in diverse ways (Ayala, 1982; Barigozzi, 1982; Giddings et al., 1989; Otte and Endler, 1989). Moreover, genetic differ-
Francisco J. Ayala is Donald Bren Professor of Biological Sciences and Ananías Escalante is a Ph.D. candidate in ecology and evolutionary biology at the University of California, Irvine. Colm O'huigin and Jan Klein are scientists at the Max-Planck-Institut för Biologie, Tübingen, Germany.
entiation between species cannot be investigated with the classical methods of genetics. With Mendelian methods, the presence of a gene is established by observing segregation in the progenies of crosses; but crosses between individuals from different species are characteristically not possible. Fortunately, the advent of molecular biology has opened up the investigation of the genetic changes associated with the speciation process. Thus, the proportion of genes that change during speciation has been ascertained in a variety of organisms (Ayala et al., 1974; Ayala, 1975; Bullini, 1982; Avise, 1994). Moreover, DNA and protein sequence information has facilitated the reconstruction of phylogenetic relationships. Yet the DNA in the gene pool of species holds a largely untapped wealth of genetic and evolutionary information. In this paper we explore DNA sequence polymorphism in current human populations in order to shed light on some aspects of human evolution, particularly on the size of human ancestral populations as they evolved from one to another species.
Speciation by Founder Effect
The theories of ''founder effect" speciation propose that speciation often occurs after a founder event or bottleneck; that is, when a new population is established by a pair or very few individuals, as may happen in the colonization of an island, or when an established population declines severely so that extremely few individuals survive, from which a population expands again (Mayr, 1954, 1963, 1982; Carson, 1968, 1975; Templeton, 1980; Carson and Templeton, 1984). These theories claim that founder events and drastic bottlenecks are associated with random genetic drift, inbreeding, and selective changes that refashion the gene pool ("genetic revolution," Mayr, 1963) and thus increase the likelihood that a new species will arise.
The prevalence of founder events (or severe bottlenecks) in speciation is a matter of great interest and acrimonious debate. Some authors argue that founder effects prevail, whereas others reject on theoretical grounds the purported genetic consequences associated with drastically reduced population numbers (Barton and Charlesworth, 1984; Giddings et al., 1989; Otte and Endler, 1989; Galiana et al., 1993).
The evolution from hominoid ancestors that lived a few million years ago to modern humans involved several transitional species, characterized by important biological changes, such as the evolution of bipedalism and a large brain. The issue arises whether these changes may have been associated with, and perhaps a consequence of, extreme population constrictions. It has been claimed, in particular, that a severe population bottleneck of only one pair or very few individuals preceded
the evolution of modern humans. This claim has been erroneously founded on the inference that the mitochondrial DNA polymorphisms of modern humans can be traced to a single woman who lived some 200,000 years ago (Cann et al., 1987; Stoneking et al., 1990; Vigilant et al., 1991). As we shall show, DNA polymorphisms in the major histocompatibility complex (MHC) of humans and other primates manifest that no severe population bottleneck has occurred in human evolution.
The Major Histocompatibility Complex
The MHC is an array of genetic loci that specify molecules with a major role in tissue compatibility and defense against foreign substances. The MHC is present in all mammals, birds, amphibians, and fishes, and it may in fact exist in all vertebrates (Klein, 1986; Bjorkman and Parham, 1990). MHC molecules present on the surfaces of certain cells bind fragments of proteins (antigens) and present them to thymus-derived lymphocytes (T cells) expressing T-cell receptors on their surfaces. The clone of T lymphocytes bearing receptors that match a particular combination of protein fragment and MHC molecule is stimulated, by the contact with the antigen-presenting cells, to proliferate and to initiate the specific arm of the immune response, including the secretion of specific antibodies. The MHC molecules thus protect against pathogens and parasites in general.
The recognition of protein fragments is mediated by a specialized groove on the surface of the MHC molecule, the so-called peptide-binding region (PBR) composed of some 50 amino acid residues (Bjorkman et al., 1987a, b; Brown et al., 1993). The composition of the amino acids in the PBR varies from one MHC molecule to another, and it is primarily this variation that is responsible for the tremendous polymorphism characteristic of the MHC molecules and their encoding genes. In people, as well as in some mammalian species (e.g., the house mouse), scores of alleles may exist at any one of several MHC loci, and some of the allelic pairs may differ by more than 100 nucleotide substitutions (Klein and Figueroa, 1986; Marsh and Bodmer, 1991, 1993; Bontrop, 1994).
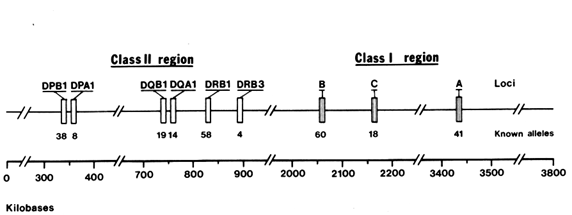
The human MHC, also referred to as the human leukocyte antigen (HLA ) complex, consists of about 100 genes located on chromosome 6, extending over a DNA region that is four million nucleotide pairs in length. The MHC genes fall into two distinct classes, I and II, separated by a set of genes with functions mostly unrelated to the immune response (Figure 1).
The origins of the MHC polymorphisms have been studied extensively in rodents (Arden and Klein, 1982; Figueroa et al., 1988; McCon-
nell et al., 1988) and primates (Mayer et al., 1988; Lawlor et al. , 1988; Fan et al., 1989; Gyllenstein and Erlich, 1989; and Gyllenstein et al., 1990). In both orders, convincing evidence has been obtained indicating that some of the allelic lineages at the MHC loci are several million years (Myr) old. We will review the allelic diversity found at one particular HLA class II locus, the DRB1 gene. Length constrains us from reporting other HLA loci, such as DPB1 and DQB1 (see Figure 1), that lead to similar conclusions.
Trans-Specific Polymorphisms
We are interested in identifying trans-specific polymorphisms, that is, sets of allelic lineages that may have been passed from an ancestral species to its descendant species. Two ways are available for detecting trans-specific polymorphisms. The first method seeks to identify alleles in a species that are more different from each other than they are from alleles in a separate species. Consider a situation in which there are two alleles, 01 and 02, at a locus in species Z, which are both passed on to descendant species X and Y (Figure 2). In the new species the two alleles accumulate mutations, turning into alleles 0101, 0102, 0103, etc. and 0201, 0202, 0203, etc. which are different from one another and from the original alleles 01 and 02. If we compare these alleles, we find that some alleles of the same species, such as 0101 and 0201, are more different from each other than some alleles from different species, such as 0101 and 0103. If we now draw a tree depicting the descent of these alleles, it will not coincide with the phylogenetic tree of the species: the 01 and 02 alleles split from a common ancestor before the two species X and Y evolved from their common ancestral species Z.
An example of trans-species polymorphism is depicted in Figure 3. Here, two human alleles at the DRB1 locus (HLA DRB1*0302 and *0701 ) differ in the exon encoding the PBR by 31 nucleotide substitutions, whereas one of them (HLA DRB1*0302) differs from the corresponding chimpanzee allele (Patr DRB1*0305) by 13 substitutions, and the other (HLA DRB1*0701) differs from the corresponding chimpanzee allele (Patr DRB1*0702) by 2 substitutions only. In terms of genetic distance (the number of nucleotide differences divided by the total number of sites compared), each of the two human alleles is more closely related to a chimpanzee allele than the two human alleles are to each other. This relationship is reflected in a family tree of the four alleles, which shows that the two human alleles diverged from a common ancestral gene before the ancestors of the human and chimpanzee species separated from each other around 6 Myr ago.
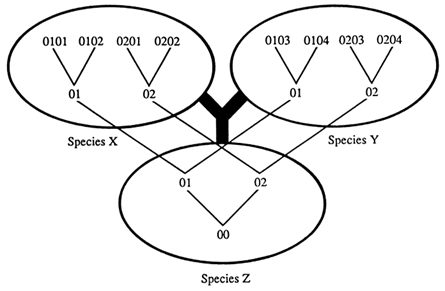
Figure 2 Phylogeny of species (thick lines) and of alleles (thin lines). Numbers indicate progressively diverging alleles at a given locus. The species and allele trees may not coincide whenever a polymorphism is passed from species Z on to species X and Y; i.e., the root of the alleles may be deeper than species Z. Some alleles, such as 0101 and 0201, of the same species are more different from each other than some alleles, such as 0101 and 0103, of different species.
The second method for detecting trans-species polymorphism depends on knowing the rate of evolution (i.e., rate of nucleotide substitution) for the particular gene. Genetic distances between alleles can
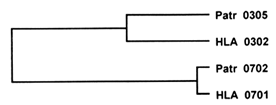
Figure 3 Trans-species polymorphism manifested by the genealogy of four alleles at the MHC DRB1 locus in humans (HLA) and chimpanzee (Pan troglodytes, Patr). The two HLA allelic lineages predate the divergence of the human and chimpanzee lineages. The lengths of the horizontal branches are roughly proportional to the genetic distance, or amount of genetic change, separating the alleles.
then be transformed into times since divergence from the ancestral allele (Figure 4). (In reality, the two methods are equivalent. Thus, if 80 DRB1 alleles sequenced in other primates were incorporated in Figure 4 with the 58 human alleles, we would see that many human alleles are more closely related to alleles from other primates than to one another. One practical reason not to represent all 138 alleles here is the enormous size of the required figure.)
Evolutionary History of the DRB1 Gene Locus
Exon 2 of the DRB1 gene consists of 270 nucleotides that specify all the b-chain amino acids involved in peptide binding. No fewer than 58 distinct human alleles have been identified that differ in their exon 2 nucleotide sequences. Figure 4 is a phylogenetic reconstruction of the 58 alleles.
The time scale has been determined by the "minimum-minimum" method (Satta et al., 1991, 1993), which is based on the comparison between pairs of species that share the same divergence node, such as the three pairs orangutan–human, orangutan–gorilla, and orangutan–chimpanzee. The minimum distance observed in such a set of comparisons will correspond to alleles that diverge at, or close to, the time of the species divergence. (Alleles that were polymorphic at the time of the species divergence will show larger distances than the minimum-minimum value.) A plot of minimum-minimum distances versus the correspondent divergence times gives an estimate of rate of evolution.
As can be seen in Figure 4, all 58 DRB1 alleles have persisted through the last 500,000 years, but coalesce into 44 ancestral lineages by 1.7 Myr before present (B.P.), 21 lineages by 6 Myr B.P., and 10 lineages by 13 Myr B.P. It is apparent that the DRB1 polymorphism is ancient, so that numerous alleles have persisted through successive speciation events. In order to pass 58 alleles through the generations, no fewer than 29 individuals are required at any time. As we shall presently see, the size of the human populations needs to be much greater than that in order to retain the DRB1 polymorphisms over the last few million years.
Persistence of Polymorphisms
What are the conditions under which we can expect a trans-specific passage of gene polymorphisms? There are several factors that determine the fate of gene polymorphisms during speciation. We will consider three of them: the time (t) elapsed since the divergence of the species, the selective value (s) of the alleles, and the size (N) of the gene pool.

Figure 4 Evolutionary tree (genealogy) of 58 HLA DRB1 alleles. The tree is based on the DNA sequence of exon 2, which consists of 270 nucleotides. Genetic distances are estimated by Kimura's (1983) two-parameter method (with pairwise elimination of any unsequenced sites). The tree was obtained with a standard UPGMA algorithm. The rate of nucleotide substitution, estimated by the minimum-minimum method (Satta et al., 1991, 1993) for exon 2, is 2 × 10-9 per site, per year. As divergence times from the human lineage we use: chimpanzee and gorilla, 6 My; orangutan, 13 My; Old-World monkeys, 25 My; New-World monkeys, 40 My; prosimians, 65 My. Satta et al. (1993) have estimated rates of 1.84 × 10-9 for class I genes and 1.18 × 10-9 for DRB1, estimates that would increase the range of the time scale at bottom to 33 My and 51 My, respectively. Sequence data are mostly from Marsh and Bodmer (1993).
Alleles can have positive, negative, or no selective value relative to preexisting alleles. An allele with a negative selective value is usually eliminated from the gene pool soon after its appearance and does not contribute to polymorphism.
The fate of an allele with no selective value (a neutral allele) depends on chance only: the allele can drift and eventually become lost or fixed (i.e., replace all other alleles at that locus). Theory says that a neutral allele that eventually becomes fixed takes on average 4N generations (with a very large standard error) from its appearance by mutation to its fixation (Kimura, 1983; Nei, 1987), where N is the effective population size (which for our purposes is roughly the number of gene pairs at a given locus that are passed on from one generation to the next). In a population with N = 106 individuals and generation time = 15 years, it will take on the average 60 Myr for a new neutral allele to become fixed; a polymorphism will persist through that time. If the generation time is 1 year, the polymorphism will persist for 4 Myr.
If a new allele 01 has a positive selective value (it has a selective advantage, s, relative to another allele, 02), the expected time to fixation is less than 4N generations and is, approximately, (2/ s)loge(2N) generations (Kimura and Ohta, 1969; Kimura, 1983; Nei, 1987). In the case of balancing selection due to overdominance (i.e., when heterozygotes have higher fitness than either homozygote) or frequency-dependent selection (i.e., if an allele is at an advantage when it is rare but becomes disadvantageous when it gets common), a polymorphism can persist without fixation for as long as the selection pressure persists (Takahata, 1990). Moreover, a balanced polymorphism will create a barrier that slows down or completely hinders the fixation of neutral mutations that are so closely linked to the positively selected site that recombination occurs between the two only rarely (Klein and Takahata, 1990).
The average duration of a species in the paleontological record is 4 Myr (Raup, 1994). Balanced as well as neutral polymorphisms may well
persist longer than it takes one species to evolve into a different species, or to diverge into two or more species.
Evolutionary Substitutions and Coalescence Theory
Now that we have defined the conditions for the passage of trans-species polymorphisms, we shall turn the argument around and attempt to extrapolate from the existing polymorphisms to the events converting an ancestral gene pool to a new pool.
The coalescence theory examines the genealogical relationships between genes (see Griffiths, 1980; Hudson, 1990). According to this theory, all alleles present in an extant pool must have descended from a single allele (to which they coalesce). For neutral alleles in a random mating population, the mean coalescence time is given by 4 N[1 - (1/i)] generations, where i is the number of sampled genes. For any two genes (i = 2), the mean coalescence time reduces to 2N generations; for a large number of genes, the mean coalescence time approaches 4N generations. These relationships are important because, if we know the time when alleles coalesce, we can estimate N, the mean number of individuals in the species.
The coalescence theory was originally developed for neutral genes (Kingman, 1982a, b; Tajima, 1983; Tavaré, 1984; and Takahata and Nei, 1985) but has been recently extended to allelic genealogies under balancing selection (Takahata, 1990, 1993a, b; Takahata and Nei, 1990). In the latter situation, the theory has the same mathematical structure as in neutral gene genealogy except for a time-scaling factor, fs. This factor is a function of population size N, selected mutation rate u, and selection coefficient s, and is given approximately by the formula

Given i sampled genes, the coalescence theory permits one to estimate the number of distinct genes, j, that existed at a certain time, with time measured in 2Nfs generations. The allelic phylogeny of the DRB1 locus (Figure 4) shows 58 HLA distinct alleles, of which 10 lineages were already in existence 13 Myr ago. The complete set coalesces 30 Myr ago, that is, before the emergence of the hominoids.
If we assume an average generation time of 15 years, the coalescence would occur two million generations ago. In the case of neutral genes, the expected time to coalescence, given by 4N generations, yields a value of N = 500,000 individuals as the long-term mean
effective size of the species. This estimate has a large variance (see below).
In the case of balanced selection, the population size required for the maintenance of a polymorphism is smaller. On the assumption of overdominance, with a heterozygote advantage on the order of 0.01, and 10-8 selected mutation rate per site per generation, the HLA polymorphism at the DRB1 locus requires a mean effective size of 100,000 individuals over the last 30 million years (Takahata et al., 1992). Computer simulations lead to estimates also on the order of 105 individuals as the long-term size of the ancestral populations leading to modern humans (Klein et al., 1990; Takahata, 1991, 1993a).
If we assume a mean population size of 105 individuals and a long-term generation time of 15 years, the expected coalescence for neutral alleles is 6 Myr, which is much less than the 30 Myr coalescence of the DRB1 alleles. Although the coalescence estimate has a large variance, it seems that either our ancestral population was even larger than 105 or, as assumed, balancing selection accounts for the long-term persistence of the MHC polymorphisms. The presence of balancing selection is supported by the analysis of the DNA sequences of HLA alleles. In codons specifying amino acids of the PBR, variation at the first and second positions is significantly higher than at the third position, and this observation is taken as evidence that positive selection acts on the first two positions (Hughes and Nei, 1988, 1992). Moreover, Hill et al. (1991, 1992a, b; see Miller, 1994) have shown that MHC polymorphism may increase resistance to Plasmodium falciparum, the parasite responsible for malignant malaria.
Estimates of the magnitude of the selection coefficient, s, that maintains the MHC polymorphisms vary from locus to locus, but range from 0.0007 to 0.019 (Satta et al., 1994). It seems unlikely that the selection coefficient would be in any case much larger than 0.01–0.03, but even large selection coefficients do not allow for the long-term persistence of polymorphisms except in the presence of large populations. For example, only 7 alleles can be maintained in a population of N = 1000, even with overdominant selection as unreasonably large as 0.3 (Figure 5; Klein et al., 1990).
Population Bottlenecks and the Origin of Modern Humans
We have concluded, assuming an overdominant selective advantage of 0.01, that the long-term effective population size of the human lineage for the last 30 Myr is 100,000 individuals or larger. When population size oscillates, smaller numbers have a large effect on the value of N, since
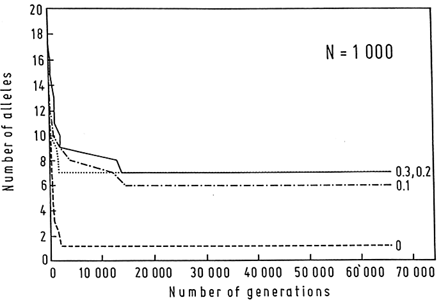
Figure 5 Computer simulation of the loss of alleles over 65,000 generations in a population consisting of 1000 individuals. Initially 20 alleles are present, each at 0.05 frequency. Mating is random and there is no mutation or migration. Heterozygotes reproduce with a higher probability s than homozygotes; s = 0.1, 0.2, and 0.3. Without selection, all alleles but one are soon eliminated. When strong overdominant selection is assumed, 6–7 alleles persist; but the magnitude of the selection is of little consequence within the range of s values used. (From Klein et al., 1990.)
the effective population size is the harmonic mean of the population size over time. Thus, at nearly all times the population must have consisted of 100,000 or more individuals. The question, nevertheless, arises whether an occasional population bottleneck may have occurred, and how small could a bottleneck be. It has been suggested that a very narrow population bottleneck occurred at the transition from archaic to modern Homo sapiens, some 200,000 years ago (see Cann et al., 1987; Stoneking et al., 1990; Vigilant et al., 1991).
The consequences of a bottleneck depend not only on the size, Nb, of the bottleneck, but also on the number, tb, of bottleneck generations. The role of tb is particularly significant in species with limited capacity for population increase. The rate of growth of human populations throughout the Pleistocene is estimated at 0.02% per generation (Spuhler, 1993, p. 279).
A useful measure for evaluating the effects of a bottleneck is the ratio
TABLE 1 Minimum population size at a bottleneck
|
|
|
|
Alleles through bottleneck |
|
|
s |
R |
N |
40/50 |
60/70 |
|
0 |
— |
— |
270–300 |
458–490 |
|
0.01 |
— |
— |
292–302 |
454–462 |
|
0 |
1.1 |
10,000 |
460–470 |
|
|
0 |
1.1 |
5,000 |
|
750–790 |
|
0 |
1.05 |
5,000 |
980–1010 |
|
|
0 |
1.01 |
10,000 |
1540–1560 |
|
|
0 |
1.01 |
5,000 |
|
2120–2180 |
|
0.01 |
1.01 |
5,000 |
|
2140–2180 |
|
The two columns on the right give the minimum number of individuals required for passing either 40 (out of 50) or 60 (out of 70) alleles, with a 95% probability. The initial bottleneck is always 10 generations. In some cases it is assumed that the population grows at a rate R per generation before reaching the equilibrium size N. The selective value due to overdominance is s. Each value is based on 300 computer simulations. |
||||
Nb/tb, which if smaller than 10 will have drastic effects in reducing genetic variation (Takahata, 1993a). Thus a bottleneck of 100 individuals would substantially reduce genetic variation if it would last 10 or more generations. Balancing selection facilitates the persistence of polymorphisms through a bottleneck. But because alleles behave as neutral whenever Ns < 1, if the selection is weak, such as s = 0.01, N has to be correspondingly large, at least 100, for selection to play a role. The persistence of HLA polymorphisms over millions of years requires that the size of human ancestral populations be at least Ns = 10 at all times (Takahata, 1990). If s = 0.01, the minimum population size possible at any time would be Nb = 1000. The minimum number must have been in fact much larger, because human population bottlenecks cannot last just a few generations, since many are required for a human population to grow from 1000 to the long-term mean of 100,000 individuals.
We have explored how small the population bottlenecks would be by computer simulations (Table 1). If a bottleneck lasts 10 generations and we ignore the time required for a population to grow back to its mean size, the smallest bottleneck that allows the persistence of 40 allelic lineages, out of 50 present before the bottleneck, with a probability of 95%, is 270–300 individuals. If the number of alleles passing through the bottleneck is 60 as in DRB1 (and assuming 70 alleles before the bottleneck), the minimum population size is 458–490. When we take into account the time required for the population to recover to its average size, the minimum population size at the bottleneck becomes substantially larger. For example, if we assume a rate of population increase of 1% per generation (which is 50 times greater
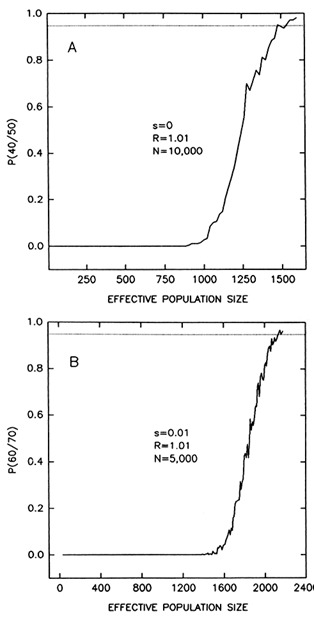
Figure 6 Probability that a number of alleles will persist after a bottleneck lasting 10 generations, as a function of population size at the bottleneck. The graphs represent averages of 300 computer simulations. (A) Probability that 40 alleles will persist out of 50 present at the start of the bottleneck; alleles are neutral (s = 0) and the population grows at a rate R = 1.01 per generation until it reaches a size N = 10,000 individuals. (B) Probability that 60 alleles will persist out of 70 present at the start of the bottleneck; heterozygotes have 1% fitness advantage over homozygotes (s = 0.01); R = 1.01; N = 5000. A probability of 95% requires minimum population sizes of 1540-1600 (A) and 2140-2180 (B) individuals. The probability increases sharply with population size and, thus, the minimum size is only moderately reduced for probabilities less than 0.95.
than the average rate of growth of human populations throughout the Pleistocene; Spuhler, 1993, p. 279), a minimum effective population size of 2120-2180 individuals is required for passing 60 out of 70 alleles. (Recovery is simulated only to a population size of 5000 individuals in this case; recovery to 100,000 individuals will further increase the minimum population size.) Selection has very little effect under these conditions (compare the two top rows or the two bottom rows in Table 1). Reducing the probability of passing the polymorphisms to some value smaller than 95% is also of little consequence (Figure 6).
The conclusion follows that human ancestral populations could never have been smaller than two or three thousand individuals at any time over the last several million years. This conclusion might be strengthened by taking into account that extensive polymorphisms exist at other HLA loci (and at other genes as well). For example, 63 primate alleles are known of the DQB1 gene, 17 of them in humans. As many as 14 DQB1 human alleles predate the origin of Homo erectus, 9 alleles predate the divergence of the human and chimpanzee lineages, and 7 alleles predate the divergence of the human and orangutan lineages. Another instance is the occurrence of multiallelic polymorphisms in the b-globin family that yield at least 17 haplotypes, the coalescence of which goes back to 450,000 years B.P. or earlier (Fullerton et al., 1994) and would be consistent with an effective population of 10,000 individuals through that time span.
Theories Of Human Origins
The origin of anatomically modern humans, Homo sapiens sapiens, occurred around 200,000 years B.P. The transition from H. erectus to H. sapiens happened around 500,000 years B.P., and the emergence of H. erectus occurred in Africa somewhat earlier than 1.7 Myr B.P. (Swisher et al., 1994, Gibbons, 1994). The hominid lineage diverged from the chimpanzee lineage about 6 Myr B.P., and the orangutan lineage
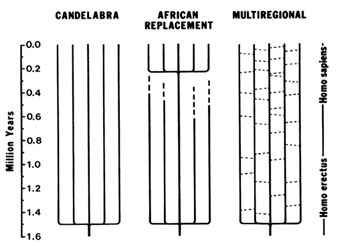
Figure 7 Three models of human evolution during the Pleistocene. The candelabra model proposes that the transition from H. erectus to H. sapiens occurred independently in different parts of the World. The African replacement model proposes that populations of H. erectus and archaic H. sapiens became extinct in Asia and Europe and were replaced by anatomically modern humans immigrating from Africa. The multiregional model proposes regional continuity and local selection pressures in different parts of the world, but with gene flow (indicated by the dashes connecting the vertical lines that represent different regional populations) due to occasional migrations between populations.
diverged from the lineage of African apes and humans about 13 Myr B.P.
H. erectus spread out of Africa shortly after its emergence from Homo habilis. Fossil remains of H. erectus are known from Indonesia (Java), China, and the Middle East, as well as Africa (Gibbons, 1994). H. erectus fossils from Java have been dated 1.81 ± 0.01 and 1.66 ± 0.04 Myr B.P. (Swisher et al., 1994). There are three models concerning the geographic origin of modern humans: candelabra, African replacement, and multiregional (Figure 7).
The candelabra model was proposed by Carleton S. Coon (1962), who argued that ''Homo erectus evolved into Homo sapiens not once but five times, as each subspecies, living in its own territory, passed a critical threshold from a more brutal to a more sapient state." Moreover, the threshold was crossed at different times, the Caucasoid race becoming sapiens first, whereas the Congoloids reached the sapiens condition some 200,000 years later. Coon's proposal was effectively criticized by several authors who pointed out the impossibility that the same species would
evolve independently twice, let alone five times, in different regions and at different times (Dobzhansky, 1963). The candelabra model is, moreover, contradicted by fossil (Aiello, 1993; Spuhler, 1993) and molecular (Takahata, 1993a, pp. 13, 19) evidence.
The African replacement model proposes that modern humans first arose in Africa somewhat prior to 100,000 years B.P. and from there spread throughout the world, replacing elsewhere the preexisting populations of H. erectus or archaic H. sapiens (Stringer, 1990, 1992; Stringer and Andrews, 1988). One extreme version of this hypothesis (the Noah's Ark model) proposes that the transition of archaic to modern H. sapiens was associated with a very narrow bottleneck, consisting of only two or very few individuals who are the ancestors of all modern mankind.
The Noah's Ark model is supported by an interpretation of mitochondrial DNA analysis showing that the diverse mitochondrial DNA sequences found in modern humans coalesce to one ancestral sequence, the "mitochondrial Eve" or "mother of us all," that existed in Africa about 200,000 years ago (Cann et al., 1987; Stoneking et al., 1990; Vigilant et al., 1991). This conclusion has been challenged on grounds concerning (1) whether the coalescence is to Africa, (2) the time of the coalescence, and (3) the inference of a population bottleneck (e.g., Templeton, 1992). The actual date of coalescence depends on assumptions about evolutionary rates. Based on a time of divergence between humans and chimpanzees of 6 Myr, the time to coalescence for mitochondrial DNA polymorphism has been recently estimated at 298,000 years B.P., with a 95% confidence interval of 129,000–536,000 years (Ruvolo et al., 1993).
The inference that a narrow bottleneck occurred at the time of the coalescence is based on a confusion between gene genealogies and individual genealogies. Gene genealogies gradually coalesce towards a unique DNA ancestral sequence, whereas individual genealogies increase by a factor of 2 per generation: an individual has two parents in the previous generation, four ancestors in the generation before that, and so on. (The theoretical number of ancestors for any one individual becomes enormous after some tens of generations, but "inbreeding" occurs: after some generations, ancestors appear more than once in the genealogy.)
As we pointed out above, assuming an effective population of N individuals, mean coalescence is 4N generations for nuclear polymorphisms. Mitochondrial DNA is haploid and maternally inherited; hence, the mean coalescence is 2Nf, where Nf is the number of mothers. If we assume 20 years for a human generation throughout the Pleistocene, 298,000 years to coalescence implies an effective mean population size of 7450 mothers or an effective population size of 14,900 humans. The 95% confidence time estimate of 129,000–536,000 years yields 6,450–26,800
generations, corresponding to a mean effective population size of 6,450–26,800 individuals.
There is one more factor to take into account in the calculations just made. We have used mean estimates to coalescence time, but these estimates have large variances. When the sample of genes is large, the standard deviation of the mean for nuclear genes is larger than 2N (Nei, 1987, eq. 13.74); for mitochondrial DNA it is larger than N/2. The 95% confidence interval for the number of generations to coalescence will correspondingly extend at the upper end to more than 2N (53,600) generations for mitochondrial DNA.
Thus the mitochondrial DNA sequence data are consistent with a mean effective population size between somewhat less than 10,000 and more than 50,000 individuals throughout the Pleistocene. This population size is, in turn, consistent with the estimate based on the HLA DRB1 polymorphism of a mean population size of 100,000 individuals for human ancestors over the last 30–40 Myr. The mitochondrial DNA data are also consistent with the result that no bottleneck smaller than several thousand individuals could have happened in hominid history.
The multiregional model emphasizes regional continuity in the evolution from H. erectus to archaic H. sapiens, and later to anatomically modern humans. Several variations of this model have been formulated (Aiello, 1993) that differ in the relative role given to two factors: (i) the prevalence of Africa in the gradual emergence of modern features, and (ii) the amount of migration between regions. At one extreme, the "African hybridization and replacement model" proposes that modern humans first evolved in Africa, but their dispersal throughout the world was accompanied by certain amount of hybridization with indigenous premodern populations (Braüer, 1984, 1992). A variant that might be called the "Levantine hybridization and replacement model" is statistically supported by a correlation analysis of morphological traits that favors the Middle East over Africa as the origin of modern human traits (Waddle, 1994). At the other extreme, some authors deny a recent African origin for modern humans while emphasizing regional continuity and gene exchange throughout Africa, Europe, and Asia (Wolpoff et al., 1988; Wolpoff, 1989). Other authors have proposed an intermediate "assimilation model'' that gives preponderance to Africa in the emergence of anatomically modern traits but attributes major importance to gene flow and differential selection pressures resulting in morphological differentiation among regions (Smith et al., 1989; Smith, 1992; Spuhler, 1993).
The paleontological data favor some form of the multiregional model, but one that gives preponderance to Africa (or the Levant, according to Waddle, 1994) as the locus where modern human traits first appear
(Aiello, 1993). There is evidence of regional continuity, and also of the incorporation of traits from one into another population (Aiello, 1993; Spuhler, 1993).
Molecular Evidence and the Multiregional Model
Does the molecular evidence favor any particular model of the origin of modern humans? The HLA evidence contradicts the Noah's Ark model and evinces that the ancestral population of modern humans was at no time smaller than several thousand individuals, a result also consistent with the mitochondrial DNA data.
Two recent molecular studies favor some degree of regional continuity over complete African replacement. The first study concerns polymorphisms in the genes for green and red visual pigments. Color vision in animals is mediated by light-sensitive pigments consisting of a chromophore covalently linked to a protein moiety (opsin). The genes coding for opsins in the red and green pigments are located on the long arm of chromosome X, whereas the one for the blue pigment is on chromosome 7 (Deeb et al., 1994). In humans, the red and green opsin genes are highly homologous and consist of six exons. The duplication of these two genes has been dated to 30–40 Myr B.P., shortly after the divergence of the Old- and New-World primates.
The green and red opsin genes have now been sequenced in a sample of 16 chimpanzees, 7 gorillas, and 4 orangutans, yielding a total of 14 biallelic polymorphic sites (all in either exon 2 or 4) (Deeb et al., 1994). Six of these polymorphisms are also found in humans, which indicates that they are of ancient origin predating the divergence of humans and apes.
One of these trans-specific polymorphisms involves the amino acid residue 65, which in the green opsin gene of orangutans and humans can be either threonine or isoleucine (Deeb et al., 1994). The relevant result is that this polymorphism has been found in Caucasians (the Ile-65 allele in 4 out of 120 individuals) but not in a sample of 56 individuals of African ancestry and 49 of Asian ancestry (Deeb et al., 1994). It is possible that the Ile-65 allele may eventually be found in African populations. It is also possible that it may have been lost from African and Asian populations in recent times, that is, after the emergence of modern humans. But since this polymorphism is millions of years old, loss of the allele over the long period since the migration of H. erectus out of Africa is more likely than a recent loss. In the replacement model, migrants from Africa colonize other parts of the world and replace any preexisting populations within the last 200,000 years. It would seem unlikely that the polymorphism would have been passed on to Caucasian populations and become thereafter lost in the
larger African population. Thus, the opsin polymorphism argues (mildly) against a complete replacement of the Caucasian gene pool by African populations.
The second example concerns an autosomal recessive disorder in lipid metabolism due to the absence of apolipoprotein C-II, the physiological activator of lipoprotein lipase, a key enzyme in very low density lipoprotein metabolism. Two deleterious alleles, one from a Venezuelan Caucasian family and one from a Japanese family, share a frameshift mutation suggesting common ancestry (Li et al., 1993). These two mutants diverged from the normal allele at least 2 Myr B.P. (Li et al., 1993). The persistence of two defective alleles over such a long time is a puzzle, perhaps a consequence of small heterozygote advantage. But this persistence (i) argues against extremely small population bottlenecks throughout Pleistocene human history, and (ii) favors the conclusion that European and Asian H. erectus have contributed to the gene pool of modern H. sapiens (Li et al., 1993).
Summary
The major histocompatibility complex (MHC) plays a cardinal role in the defense of vertebrates against parasites and other pathogens. In some genes there are extensive and ancient polymorphisms that have passed from ancestral to descendant species and are shared among contemporary species. The polymorphism at the DRB1 locus, represented by 58 known alleles in humans, has existed for at least 30 million years and is shared by humans, apes, and other primates. The coalescence theory of population genetics leads to the conclusion that the DRB1 polymorphism requires that the population ancestral to modern humans has maintained a mean effective size of 100,000 individuals over the 30-million-year persistence of this polymorphism. We explore the possibility of occasional population bottlenecks and conclude that the ancestral population could not have at any time consisted of fewer than several thousand individuals.
The MHC polymorphisms exclude the theory claiming, on the basis of mitochondrial DNA polymorphisms, that a constriction down to one or few women occurred in Africa, at the transition from archaic to anatomically modern humans, some 200,000 years ago. The data are consistent with, but do not provide specific support for, the claim that human populations throughout the World were at that time replaced by populations migrating from Africa. The MHC and other molecular polymorphisms are consistent with a "multiregional" theory of Pleistocene human evolution that proposes regional continuity of human populations since the time of migrations of Homo erectus to the present,
with distinctive regional selective pressures and occasional migrations between populations.
We are grateful to Drs. W. M. Fitch, R. R. Hudson, and Y. Satta for reading the manuscript and offering suggestions, and to Dr. E. Barrio for help with the figures. Research in molecular evolution by the senior author is supported by National Institutes of Health Grant GM42397. A.E. is supported by a fellowship from the Banco Interamericano de Desarrollo–Consejo Nacional de Investigaciones Científicas y Tecnológicas Program, Venezuela.
References
Aiello, L. C. (1993) The fossil evidence for modern human origins in Africa: A revised view. Am. Anthropol. 95, 73–96.
Arden, B. & Klein, J. (1982) Biochemical comparison of major histocompatibility complex molecules from different subspecies of Mus musculus: Evidence for trans-specific evolution of alleles. Proc. Natl. Acad. Sci. USA 79, 2342–2346.
Avise, J. C. (1994) Molecular Markers, Natural History and Evolution (Chapman & Hall, New York).
Ayala, F. J. (1975) Genetic differentiation during the speciation process. Evol. Biol. 8, 1–78.
Ayala, F. J. (1982) Population and Evolutionary Genetics (Benjamin/Cummings, Menlo Park, CA).
Ayala, F. J., Tracey, M. L., Barr, L. G. & Ehrenfeld, J. G. (1974) Genetic and reproductive differentiation of Drosophila equinoxialis caribbensis. Evolution 28, 24–41.
Barigozzi, C. (1982) Mechanisms of Speciation (Liss, New York).
Barton, N. H. & Charlesworth, B. (1984) Founder effects, genetic revolutions, and speciation. Annu. Rev. Ecol. Syst. 15, 133–164.
Bjorkman, P. J., Saper, M. A., Samraoui, B., Bennett, W. S., Strominger, J. L. & Wiley, D. C. (1987a) Structure of the human class I histocompatibility antigen, HLA-A2. Nature (London) 329, 506–512.
Bjorkman, P. J., Saper, M. A., Samraoui, B., Bennett, W. S., Strominger, J. L. & Wiley, D. C. (1987b) The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature (London) 329, 512–521.
Bjorkman, P. J. & Parham, P. (1990) Structure, function, and diversity of class-I major histocompatibility complex molecules Annu. Rev. Biochem. 59, 253–288.
Bontrop, R. E. (1994) Nonhuman primate Mhc-DQA and -DQB second exon nucleotide sequences: a compilation. Immunogenetics 39, 81–92.
Braüer, G. (1984) A craniological approach to the origin of anatomically modern Homo sapiens in Africa and implications for the appearance of modern Europeans. In The Origins of Modern Humans: A World Survey of the Fossil Evidence, eds. Smith, F. H. & Spencer, F. (Liss, New York), pp. 327–410.
Braüer, G. (1992) Africa's place in the evolution of Home sapiens. In Continuity or Replacement? Controversies in Homo sapiens Evolution , eds. Braüer, G. & Smith, F. H. (Balkema, Rotterdam, The Netherlands), pp. 83–98.
Brown, J. H., Jardetzky, T. S., Gorga, J. C., Stern, L. J., Urban, R. G., Strominger, J. L. & Wiley, D. C. (1993) Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature (London) 364, 33–39.
Bullini, L. (1982) Ecological and ethological aspects of the speciation process. In Mechanisms of Speciation, ed. Barigozzi, C. (Liss, New York), pp. 241–264.
Cann, R. L., Stoneking, M. & Wilson, A. C. (1987) Mitochondrial DNA and human evolution. Nature (London) 325, 31–36.
Carson, H. L. (1968) The population flush and its genetic consequences. In Population Biology and Evolution, ed. Lewontin, R. C. (Syracuse Univ. Press, New York), pp. 123–127.
Carson, H. L. (1975) The genetics of speciation at the diploid level. Am. Nat. 109, 83–92.
Carson, H. L. & Templeton, A. R. (1984) Genetic revolution in relation to speciation phenomena: The founding of new populations. Annu. Rev. Ecol. Syst. 15, 97–131.
Coon, C. S. (1962) The Origin of Races (Knopf, New York).
Deeb, S. S., Jørgensen, A. L., Battisti, L., Iwasaki, L. & Motulsky, A. G. (1994) Sequence divergence of the red and green visual pigments in the great apes and man. Proc. Natl. Acad. Sci. USA 91, 7262–7266.
Dobzhansky, T. (1963) A debatable account of the origin of races. Sci. Am. 208 (2), 169–172.
Fan, W., Kasahara, M., Gutknecht, J., Klein, D., Mayer, W. E., Jonker, M. & Klein, J. (1989) Shared class II MHC polymorphism between humans and chimpanzees. Hum. Immunol. 26, 107–121.
Figueroa, F., Günther, E. & Klein, J. (1988) MHC polymorphism predating speciation. Nature (London) 335, 265–267.
Fullerton, S. M., Harding, R. M., Boyce, A. J. & Clegg, J. B. (1994) Molecular and population genetic analysis of allelic sequence diversity at the human b-globin locus. Proc. Natl. Acad. Sci. USA 91, 1805–1809.
Galiana, A., Moya, A. & Ayala, F. J. (1993) Founder-flush speciation in Drosophila pseudoobscura: A large-scale experiment. Evolution 47, 432–444.
Gibbons, A. (1994) Rewriting—and redating—prehistory. Science 263, 1087–1088.
Giddings, L. V., Kaneshiro, K. Y. & Anderson, W. W. (1989) Genetics, Speciation, and the Founder Principle (Oxford Univ. Press, New York).
Griffiths, R. C. (1980) Lines of decent in the diffusion approximation of neutral Wright-Fisher models. Theor. Popul. Biol. 17, 37–50.
Gyllenstein, U. B. & Erlich, H. A. (1989) Ancient roots for polymorphism at the HLA-DQa locus in primates . Proc. Natl. Acad. Sci. USA 86, 9986–9990.
Gyllenstein, U. B., Lashkari, D. & Erlich, H. A. (1990) Allelic diversification at the class II DQB locus of the mammalian major histocompatibility complex. Proc. Natl. Acad. Sci. USA 87, 1835–1839.
Hill, A. V. S., Allsopp, C. E. M., Kwiatowski, D., Anstey, N. M., Twumasi, P., Rowe, P. A., Bennett, S., Brewster, D., McMichael, A. J. & Greenwood, B. M. (1991) Common West African HLA antigens are associated with protection from severe malaria. Nature (London) 352, 595–600.
Hill, A. V. S., Kwiatkowski, D., McMichael, A. J., Greenwood, B. M. & Bennett, S. (1992a) Maintenance of MHC polymorphism. Nature (London) 355, 402–403.
Hill, A. V. S., Elvin, J., Willis, A. C., Aidoo, M., Alsopp, E. E. M., Gotch, F. M., Gao, X. M., Takiguchi, M., Greenwood, B. M., Townsend, A. R. M. , McMichael, A. J. & Whittle, H. C. (1992b) Molecular analysis of the association of HLA-B53 and resistance to severe malaria. Nature (London) 360, 434–439.
Hudson, R. R. (1990) Gene genealogies and the coalescent process. Oxford Surv. Evol. Biol. 7, 1–44.
Hughes, A. L. & Nei, M. (1988) Pattern of nucleotide substitution at major histo-
compatibility complex class I loci reveals overdominant selection. Nature (London) 335, 167–170.
Hughes, A. L. & Nei, M. (1992) Maintenance of MHC polymorphism. Nature (London) 355, 402–403.
Kimura, M. (1983) The Neutral Theory of Molecular Evolution (Cambridge Univ. Press, Cambridge, U.K.).
Kimura, M. & Ohta, T. (1969) The average number of generations until fixation of a mutant gene in a finite population. Genetics 61, 763–771.
Kingman, J. F. C. (1982a) The coalescent. Stochast. Processes Appl. 13, 235–248.
Kingman, J. F. C. (1982b) On the genealogy of large populations. J. Appl. Probab. 19, 27–43.
Klein, J. (1986) The Natural History of the Major Histocompatibility Complex (Wiley, New York).
Klein, J. & Figueroa, F. (1986) Evolution of the Major Histocompatibility Complex. CRC Crit. Rev. Immunol. 6, 295–386.
Klein, J., Gutknecht, J. & Fischer, N. (1990) The major histocompatibility complex and human evolution. Trends Genet. 6, 7–11.
Klein, J. & Takahata, N. (1990) The major histocompatibility complex and the quest for origins. Immunol. Rev. 113, 5–25.
Lawlor, D. A., Ward, F. E., Ennis, P. D., Jackson, A. P. & Parham, P. (1988) HLA-A and B polymorphisms predate the divergence of humans and chimpanzees. Nature (London) 355, 268–271.
Li, W.-H., Xiong, W., Liu, S. A.-W. & Chan, L. (1993) Nucleotide diversity in humans and evidence for the absence of a severe bottleneck during human evolution. In Genetics of Cellular, Individual, Family, and Population Variability, eds. Sing, C. F. & Hanis, C. L. (Oxford Univ. Press, New York), pp. 253–261.
Marsh, S. G. E. & Bodmer, J. G. (1991) HLA class II nucleotide sequences, 1991. Immunogenetics 33, 321–334.
Marsh, S. G. E. & Bodmer, J. G. (1993) HLA class II nucleotide sequences, 1992. Immunogenetics 37, 79–94.
Mayer, W. E., Jonker, M., Klein, D., Ivanyi, P., van Seventer, G. & Klein, J. (1988) Nucleotide sequences of chimpanzee MHC class I alleles: evidence for trans-species mode of evolution. EMBO J. 7, 2765–2774.
Mayr, E. (1954) Change of genetic environment and evolution. In Evolution as a Process, eds. Huxley, J., Hardy, A. C. & Ford, E. B. (Allen & Unwin, London), pp. 157–180.
Mayr, E. (1963) Animal Species and Evolution (Harvard Univ. Press, Cambridge, MA).
Mayr, E. (1982) Processes of speciation in animals. In Mechanisms of Speciation, ed. Barigozzi, C. (Liss, New York), pp. 1–19.
McConnell, T. J., Talbot, W. S., McIndoe, R. A. & Wakeland, E. K. (1988) The origin of MHC class II gene polymorphism within the genus Mus. Nature (London) 332, 651–654.
Miller, L. H. (1994) Impact of malaria on genetic polymorphism and genetic diseases in Africans and African Americans. Proc. Natl. Acad. Sci. USA 91, 2415–2419.
Nei, M. (1987) Molecular Evolutionary Genetics (Columbia Univ. Press, New York).
Otte, D. & Endler, J. A. (1989) Speciation and Its Consequences (Sinauer, Sunderland, MA).
Raup, D. M. (1994) The Role of Extinction in Evolution. Proc. Natl. Acad. Sci. USA 91, 6758–6763.
Ruvolo, M., Zehr, S., von Dornum, M., Pan, D., Chang, B. & Lin, J. (1993)
Mitochondrial COII sequences and modern human origins. Mol. Biol. Evol. 10, 1115–1135.
Satta, Y., Takahata, N., Schönbach, C., Gutknecht, J. & Klein, J. (1991) Calibrating evolutionary rates at major histocompatibility complex loci. In Molecular Evolution of the Major Histocompatibility Complex, eds. Klein, J. & Klein, D. (Springer, Heidelberg), pp. 51–62.
Satta, Y., O'hUigin, C., Takahata, N. & Klein, J. (1993) The synonymous substitution rate of the major histocompatibility complex loci in primates. Proc. Natl. Acad. Sci. USA 90, 7480–7484.
Satta, Y., O'huigin, C., Takahata, N. & Klein, J. (1994) Intensity of natural selection at the major histocompatibility complex loci. Proc. Natl. Acad. Sci. USA 91, 7184–7188.
Smith, F. H. (1992) The role of continuity in modern human origins. In Continuity or Replacement? Controversies in Homo sapiens Evolution , eds. Braüer, G. & Smith, F. H. (Balkema, Rotterdam, The Netherlands), pp. 145–156.
Smith, F. H., Falsetti, A. B. & Donnelly, S. M. (1989) Modern human origins. Years. Phys. Anthropol. 32, 35–68.
Spuhler, J. N. (1993) Population genetics and evolution in the genus Homo in the last two million years. In Genetics of Cellular, Individual, Family, and Population Variability, eds. Sing, C. F. & Hanis, C. L. (Oxford Univ. Press, New York), pp. 262–297.
Stoneking, M., Jorde, L. B., Bhatia, K. & Wilson, A. C. (1990) Geographic variation of human mitochondrial DNA from Papua New Guinea. Genetics 124, 717–733.
Stringer, C. B. (1990) The emergence of modern humans. Sci. Am. 263 (6), 98–104.
Stringer, C. B. (1992) Reconstructing recent human evolution. Philos. Trans. R. Soc. London B 337, 217–224.
Stringer, C. B. & Andrews, P. (1988) Genetic and fossil evidence for the origin of modern humans . Science 239, 1263–1268.
Swisher, C. C., III, Curtis, G. H., Jacob, T., Getty, A. G., Suprijo, A. & Widiasmoro (1994) Age of the earliest known hominids in Java, Indonesia. Science 263, 1118–1121.
Tajima, F. (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics 105, 437–460.
Takahata, N. (1990) A simple genealogical structure of strongly balanced allelic lines and trans-species evolution of polymorphism. Proc. Natl. Acad. Sci. USA 87, 2419–2423.
Takahata, N. (1991) in Molecular Evolution of the Major Histocompatibility Complex, eds. Klein, J. & Klein, D. (Springer, Heidelberg), pp. 29–49.
Takahata, N. (1993a) Allelic generalogy and human evolution. Mol. Biol. Evol. 10, 2–22.
Takahata, N. (1993b) Evolutionary genetics of human paleopopulations. In: Mechanisms of Molecular Evolution, eds. Takahata, N. & Clark, A. G. (Japan Scientific Societies Press, Tokyo and Sinauer Associates, Inc., Sunderland, MA), pp. 1–21.
Takahata, N. & Nei, M. (1985) Gene genealogy and variance of interpopulational nucleotide differences. Genetics 110, 325–344.
Takahata, N. & Nei, M. (1990) Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics 124, 967–978.
Takahata, N., Satta, Y. & Klein, J. (1992) Polymorphism and balancing selection at major histocompatibility complex loci. Genetics 130, 925–938.
Tavaré, S. (1984) Line-of-descent and genealogical processes, and their applications in population genetics models. Theor. Popul. Biol. 26, 119–164.
Templeton, A. R. (1980) The theory of speciation via the founder principle. Genetics 94, 1011–1038.
Templeton, A. R. (1992) Human origins and analysis of mitochondrial DNA sequences. Science 255, 737.
Vigilant, L., Stoneking, M., Harpending, H., Hawkes, K. & Wilson, A. C. (1991) African populations and the evolution of human mitochondrial DNA. Science 253, 1503–1507.
Waddle, D. M. (1994) Matrix correlation tests support a single origin for modern humans. Nature (London) 368, 452–454.
Wolpoff, M. H. (1989) Multiregional evolution: The fossil alternative to Eden. In The Human Revolution, eds. Mellars, P. & Stringer, C. (Edinburgh Univ. Press, Edinburgh), pp. 62–108.
Wolpoff, M. H., Spuhler, J. N., Smith, F. H., Radovcic*, J., Pope, G., Frayer, D. W., Eckhardt, R. & Clark, G. (1988) Modern human origins. Science 241, 772–773.
| This page in the original is blank. |