
Opportunities and Challenges for Digital Twins in Biomedical Research
Proceedings of a Workshop—in Brief
The digital twin (DT) is an emerging technology that builds on the convergence of computer science, mathematics, engineering, and the life sciences. Given the multiscale nature of biological structures and their environment, biomedical DTs can represent molecules, cells, tissues, organs, systems, patients, and populations and can include aspects from across the modeling and simulation ecosystem. DTs have the potential advance biomedical research with applications for personalized medicine, pharmaceutical development, and clinical trials.
On January 30, 2023, the National Academies of Sciences, Engineering, and Medicine hosted a public, virtual workshop to discuss the definitions and taxonomy of DTs within the biomedical field, current methods and promising practices for DT development and use as various levels of complexity, key technical challenges and opportunities in the near and long term for DT development and use, and opportunities for translation of promising practices from other field and domains. Workshop panelists highlighted key challenges and opportunities for medical DTs at varying scales, including the varied visions and challenges for DTs, the trade-offs between embracing or simplifying complexity in DTs, the unique spatial and temporal considerations that arise, the diversity of models and data being used in DTs, the challenges with connecting data and models across scales, and implementation issues surrounding data privacy in DTs. The first in a three-part series, this information-gathering workshop will inform a National Academies consensus study on research gaps and future directions to advance the mathematical, statistical, and computational foundations of DTs in applications across science, medicine, engineering, and society.1
VISIONS AND CHALLENGES FOR DIGITAL TWINS
Reinhard Laubenbacher, University of Florida, the plenary session’s first speaker, described the DT as a computational model that represents a physical system; a data stream between the system and the model is used to recalibrate the model periodically so that it continues to represent the system as it changes over the lifespan of its operation. He stated that DTs are revolutionizing industry (e.g., aeronautics) with applications including forecasting and preventative maintenance. He proposed that this notion of preventive maintenance could be reflected in medical DTs, with the potential for patients to avoid unscheduled visits to the doctor. Medical DTs
__________________
1 To learn more about the study and to watch videos of the workshop presentations, see https://www.nationalacademies.org/our-work/foundational-research-gaps-and-future-directions-for-digital-twins, accessed February 10, 2023.
![]()
combine models of human biology with operational data to make predictions and optimize therapy. However, he cautioned that medical DTs are very different from those used in industry. The medical field leverages different types of data, which are often sparse. With this somewhat limited knowledge about humans, human biology has to be reverse-engineered and encoded in a model. Thus, experimental data (often from mice or tissue cultures) is frequently used in building the computational models for medical DTs. If implemented successfully, he indicated that medical DTs could eventually lead to a paradigm shift from curative to preventive medicine as well as help to personalize medical interventions, improve decision-support capabilities for clinicians, develop drugs more quickly at a lower cost, and discover personalized therapies.
Laubenbacher emphasized that because medical DTs are much more difficult to build than computational models, distinguishing appropriate use cases for a DT versus a model is critical. For example, to improve type 1 diabetes care, an artificial pancreas2 can be linked to a sensor in a patient’s bloodstream that continually measures glucose, which is connected to a closed-loop controller that determines how much insulin the patient’s pump should inject. Because this model recalibrates itself based on the patient’s needs, he noted that this device is a medical DT. To improve cardiac care, a three-dimensional image of a patient’s heart and computational fluid dynamics could be used to simulate blood flow to make predictions about the patient’s risk for a blood clot or to make decisions about bypass surgery. The classification of this technology depends on its use—if used once for decision support, it is a personalized model; if calculations for risk and medication are updated periodically, it is a medical DT.
Laubenbacher presented several challenges associated with building a medical DT: (1) identifying and solving difficult scientific problems that arise at different scales (e.g., systems biology, biophysics, the immune system); (2) addressing gaps in modeling (e.g., multiscale hybrid stochastic models, model design that facilitates updates and expansion, reusable models, and model standards); (3) developing appropriate collection modalities for patient data (e.g., noninvasive technologies and imaging capabilities); (4) developing novel forecasting methods (i.e., learning from successful hurricane forecasting); (5) developing data analytics methods for model recalibration from patient measurements; (6) training a highly educated workforce; and (7) creating appropriate funding models for individual medical DT projects from conception to prototype, and for larger infrastructure development projects.
Karissa Sanbonmatsu, Los Alamos National Laboratory, the plenary session’s second speaker, highlighted several differences between DTs and computational models, including that DTs should provide real-time information and insights and can be used across the lifetime of a specific asset for decision-making. DTs can be classified as component twins (i.e., modeling a single part of a system), asset twins (i.e., modeling multiple parts of a system), system twins (i.e., modeling the whole system), or process twins (i.e., focusing on how parts of a system work together).
Sanbonmatsu pointed out that in the field of integrative biology, computational methods are used to integrate many types of experimental data into a coherent structural model. Computational modeling in biology is conducted at many scales—for example, cellular and molecular scale (e.g., molecular dynamics simulations); organ, tumor, and microenvironment scale (e.g., Bayesian modeling); whole-human, multisystem, and population scale (e.g., epidemiological modeling); and multiple scales. Molecular simulations, in particular, are used to understand molecular machines and ion channels, to determine structural models, to design better drugs and vaccines, and to advance applications in biotechnology. She described four key challenges in the simulation of biomolecules: (1) many biological systems are inherently discrete, where phenomena at the atomistic level have important macroscopic implications for cell fate and disease; (2) length scales are approximately 10 orders of magnitude, and time scales are approximately 20 orders of magnitude; (3) information matters, especially the sequence of bases in the human genome; and (4) systems are highly
__________________
2 The website for Tandem Diabetes Care is https://www.tandemdiabetes.com, accessed February 27, 2023.
![]()
charged, which requires long-range electrostatic force calculations. Ultimately, such simulations require many time steps to be relevant; until supercomputers have the necessary capabilities, she continued, researchers will continue to leverage experimental data for multiscale integrative biology.
Discussion
Incorporating questions from workshop participants, Irene Qualters, Los Alamos National Laboratory, moderated a discussion with the plenary session speakers. She asked if standards would help to make DTs more rigorous and trustworthy in the biomedical community. Laubenbacher replied that the industrial space uses Modelica, a standard for building models of modular components that can be integrated for DTs. The Systems Biology Markup Language, which applies to differential equation models, is an established standard in the biomedical community; however, he noted that much more work remains in the development of standards overall for medical DTs to be used at scale.
Qualters wondered whether successful examples of DTs at the cellular and molecular scale exist. Sanbonmatsu responded that although several successful simulations exist and efforts are under way to develop real-time molecular models, the community does not yet have the capability to work in real time via DTs. Laubenbacher pointed out that future DTs would have to cross scales, because most drugs work at the cellular level but have effects at the tissue level. Qualters also posed a question about whether a DT could represent a population instead of only an individual—for example, a disease model that is continually updated with new trial data. Laubenbacher observed that EpiSimS, an agent-based simulation model, captures an entire city based on details that represent the population. He cautioned that all key characteristics of a population should be included in a medical DT, as sampling the parameter space is insufficient.
In response to a question, Sanbonmatsu remarked that a medical DT should track a person’s full medical history, including drug interactions and health conditions. Because all humans are different, the sequence of each person’s chromosomes would also be useful for the model. Laubenbacher added that collecting the right data (without invasive procedures for patients) and determining how those data influence parameter updates is a significant challenge in building medical DTs. Qualters inquired about the role of organs-on-a-chip technology in building higher-fidelity digital models. Sanbonmatsu noted that such technology will be critical in bridging the gap between the actual patient and the petri dish to build computational models of an organ. Laubenbacher commented that because so little can be measured directly in humans, these technologies are beneficial, as are good cell culture systems for primary tissue. He asserted that collecting information about humans without using mice and monkeys is a step forward.
EMBRACING OR SIMPLIFYING COMPLEXITY IN DIGITAL TWINS
During the first panel, workshop participants heard brief presentations on DTs at the cellular and molecular scale from the perspectives of pharmaceutical development, clinical practice, and research. The challenge of embracing or simplifying complexity in DTs was raised.
Jeffrey Sachs, Merck & Co., Inc., defined a DT as a simulation with sufficient fidelity for an intended purpose. This simulation could represent processes, objects, or physical systems; include visualization, artificial intelligence (AI), or machine learning (ML); and involve real-time sensors. He further described DTs as a type of modeling and simulation that is personalized to a person or a population.
Sachs provided an overview of how pharmaceutical companies use modeling and simulation in clinical trials and drug development. For example, to rank order the potential efficacy of a drug or vaccine, ab initio modeling is used, with input from physics and chemistry. To determine potential drug interactions, quantitative systems pharmacology and physiologically based pharmacokinetics techniques are leveraged, with model input from measurements of biological and chemical assays in experiments. To understand which drug infusion speed is best for an individual, population pharmacokinetic-pharmacodynamics and quantitative
![]()
systems pharmacology techniques are used, with model input from both large historical data sets of biological and chemical assays and data from the individual—a type of modeling often required by regulatory agencies.
Sachs highlighted several challenges related to assessing uncertainty in modeling. Data context matters, and one should be able to measure the right thing in the right way. Model complexity and identifiability as well as computability and data heterogeneity also present obstacles. Furthermore, a question remains about when summary results from one model should be used as input to the next-level model.
Mikael Benson, Karolinska Institute, observed that medication is often ineffective for patients with complex, malignant diseases, leading to both personal suffering and financial strain. Each disease might involve thousands of genes across multiple cell types, which can vary among patients with the same diagnosis. With high-resolution models to identify the optimal drug for each patient, DTs could bridge this gap between disease complexity and health care options. He described the Swedish Digital Twin Consortium’s3 approach: a DT of a single patient is created based on the integration of detailed clinical and high-resolution data. The DT is then computationally treated with thousands of drugs to determine which offers a cure, and the best drug is given to the patient (Figure 1). He mentioned that this approach has worked for animal models, patient cells, and clinical data analysis.
Benson pointed out that the complexity of the data and analysis required for medical DTs could disrupt medical practice, research, and publishing. DTs could unsettle explainable medical practice, as diagnoses that used to be based on a few blood tests and images would be based on mechanisms that involve thousands of genes. However, examples exist in which complex clinical data can be reduced to lower dimensions. Similarly, although traditional medical research is based on a detailed analysis of a limited number of variables, medical DT research is based on an analysis of thousands of genes across multiple cell types in mouse models through clinical studies. Each part of the study is interdependent, and one single part cannot be extracted for a manuscript without losing context. Each part contains highly complicated data, and results are difficult to validate with conventional methods; validation often depends on reproducibility studies. He asserted that journal editors should address the fact that current manuscript format and peer-review limitations do not accommodate the multilayered and multidisciplinary nature of DT studies.
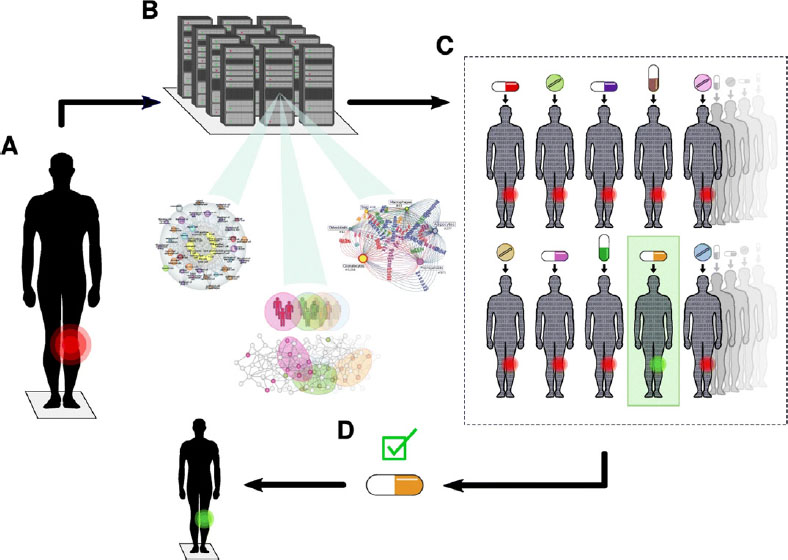
SOURCE: Mikael Benson, Karolinska Institute, presentation to the workshop, from Björnsson, B., Borrebaeck, C., Elander, N. et al., 2020, “Digital Twins to Personalize Medicine,” Genome Medicine 12:4, https://doi.org/10.1186/s13073-019-0701-3, Copyright 2019, https://creativecommons.org/licenses/by/4.0.
Juan Perilla, University of Delaware, described the study of retroviruses at the molecular level using computational models that can be interrogated to predict new properties. When this work began more than a decade ago, knowledge about cytoplasmic events was limited. Over time, those previously obscure events were revealed to be crucial in understanding how viruses take advantage of cells.
Perilla provided an overview of the process used to understand whether the HIV-1 capsid could translocate into the nucleus of a cell. To address this question, he and his team modeled biological parts and developed novel techniques to perturb the system mechanically and observed how it restored, while ensuring that the model was reproducing reality. By introducing other known chemical aspects and probing elastic properties, they could visualize the process not being captured
__________________
3 The Swedish Digital Twin Consortium website is https://sdtc.se, accessed February 27, 2023.
![]()
by experimentalists, which allowed them to make predictions, test the model, and obtain specific chemical information about the process. Understanding the system’s physical and mechanical properties confirmed that the HIV-1 capsid can translocate to the nucleus; this knowledge also enables future development of new materials.
Rommie Amaro, University of California, San Diego, described her work in developing molecular simulations at the mesoscale, with a focus on building and understanding viruses and the systems with which they interact. She and her team are creating atomic-level models of biological systems, using high-performance simulations to bring together and interrogate different types of experimental data sets and to view the trajectory of atomic-level dynamics over time.
Amaro explained that these types of simulations make it possible to predict the dynamics of structures that are otherwise invisible experimentally, thus augmenting what experimentalists cannot see (e.g., the glycan shield). Therefore, the simulations work in tandem with interrogation by experiment, she continued, and these methods enable the development of predictive models from the nanoscale to the microscale. Work is ongoing to model the dynamics of single viruses (e.g., SARS-CoV-2) using large-scale supercomputing architectures, which is crucial for vaccine development and drug design. These simulations are also being used to study airborne pathogens inside respiratory aerosols. Because DTs can be used to connect the molecular scale to population-level outcomes to make predictions, she championed Benson’s assertion that interdisciplinary peer reviewers are essential to move the field forward.
Discussion
Incorporating questions from workshop participants, Ines Thiele, National University of Ireland, Galway, moderated a discussion among the four panelists. She posed a question about strategies to validate predictions from DT simulations when experimental data cannot be obtained. Amaro explained that many indirect experiments can validate emerging predictions. When hypotheses and results can be interrogated and replicated by other research groups worldwide using the same or similar models, she asserted that trust will increase among experimentalists. Perilla suggested leveraging negative controls for validation. Sachs added that building trust relates to the application, and validation becomes especially critical in health care, where replication is only the first step in the process. Offering a clinical perspective, Benson remarked that testing a prediction in a clinical setting relates to diagnostics, sensitivity, and specificity for computational treatment with thousands of drugs. He explained that precision and recall are key for known drugs, and clinical trials for new drugs have standard measures of clinical improvement.
In response to a question about statistical considerations for DTs, Sachs commented that understanding the distribution of parameter values, characteristics, and outcomes is essential if DTs are to be used safely, as variability impacts efficacy.
Thiele asked when highly detailed precision simulations should be used instead of high-level surrogate models. Perilla explained that the study of HIV primarily focuses on cell components; the interactions of interest depend on a specific three-letter sequence and detail, which requires the incorporation of atomistic knowledge. When physical properties of a system arise from chemical properties, reducing the complexity of the system would have a negative impact. For example, although all retroviruses look alike, he pointed out that they are extremely different, and reducing the complexity of the model is dangerous. Amaro indicated that model selection depends on the domain of interest as well as what one is trying to predict. To design novel chemical matter, one has to keep track of all of the atoms in the design; however, for larger-level effects, one can coarse-grain away the most computationally intensive aspects of a calculation. She noted that over time and for particular systems, decision-making to determine what can be coarse-grained away will become more straightforward.
SPATIAL AND TEMPORAL CONSIDERATIONS IN DIGITAL TWINS
During the second panel, workshop participants heard brief presentations on DTs at the organ, tumor, and microenvironment scale in the areas of computational
![]()
oncology and ophthalmologic data sciences. A common theme emerged of addressing spatial and temporal considerations in DTs.
Tom Yankeelov, The University of Texas at Austin, emphasized the need to calibrate mechanism-based models with patient-specific data to make patient-specific predictions. He mentioned that tumor forecasts are improving in terms of accuracy, but like weather forecasts, tumor forecasts are not DTs. Promising applications for the use of DTs include linking a mechanism-based model of drug delivery to a model of tumor response, thereby enabling in silico clinical trials where the drug dose and schedule could be systematically varied. Because the optimal regimen that emerges for an individual patient is often different from the standard population-based regimen, he asserted that mechanism-based mathematical modeling is essential.
Yankeelov highlighted three key mathematical, computational, and data challenges in the development of medical DTs. First, models that capture relevant biology and that can be calibrated with available data are needed, as are models that are actionable. Second, because calibration, model selection and updates, and therapy optimization are expensive, strategies to accelerate these loops would be useful. Third, increased access to “smarter” patient data would be beneficial. As diseases and therapies become increasingly specific and complex, he urged researchers to deprioritize AI and big data in favor of mechanism-based models that can be calibrated with patient data. Furthermore, because a patient is unlikely to be cured with only one therapy, he remarked that these models should be updated continually during treatment. To begin to address these challenges, techniques from other fields could be applied such as optimal control theory, reduced order modeling, uncertainty quantification, and data assimilation.
Jayashree Kalpathy-Cramer, University of Colorado Denver, offered a relevant definition of DTs derived from ChatGPT: “a virtual representation of a physical object or system that can be used for simulation, analysis, and monitoring.”4 DTs that use medical imaging could be leveraged to inform and improve patient care via procedure planning, medical education, individual risk prediction, and prediction of cancer drug delivery.
Kalpathy-Cramer discussed a study on optimizing the regimen and dosage of anti-vascular endothelial growth factor (anti-VEGF) therapies for glioblastoma patients. The study mathematically modeled blood flow through the vessels, and three-dimensional virtual voxels generated from optical imaging in an animal model were used as input to the mathematical models. With those models, one could observe the effects of the anti-VEGF therapy on the vasculature, and what was seen in the mathematical models and simulations turned out to be similar to what was observed in humans. Presenting a completely different approach using data-driven modeling, she depicted work to identify treatment-requiring retinopathy of prematurity. ML and deep learning were used to develop a severity scale for the disease based on available data, and the trajectories were quantified of patients who would require treatment versus those who would not. A so-called DT emerged of the path an individual patient might take throughout their care, including the point at which they either respond to or require more therapy. No mathematics are used to make such predictions. However, she stressed that although data-driven approaches are powerful, they are sometimes entirely wrong. As DTs also have the potential to be both powerful and incorrect, she described a combination of data-driven and mechanistic approaches as the best path forward to make better decisions for patients.
Discussion
Incorporating questions from workshop participants, Caroline Chung, MD Anderson Cancer Center, moderated a discussion among both speakers and two experts in intelligent systems engineering and applied mathematics—James A. Glazier, Indiana University, and Petros Koumoutsakos, Harvard University, respectively. In light of the nuanced differences between computational models and DTs, Chung posed a question about how to determine whether a problem warrants the use of a DT. Yankeelov observed that if a patient has cancer and is prescribed several drugs, the order and delivery of those drugs is critical—a DT would help
__________________
4 J. Kalpathy-Cramer, University of Colorado Denver, presentation to the workshop, January 30, 2023.
![]()
determine the optimal therapeutic regimen that could be adopted in a clinical trial. Glazier added that because real-time evaluation of a drug’s effectiveness is not currently feasible, a DT of a patient could be used as a virtual control to determine whether a drug is working and to tune the therapy accordingly. Koumoutsakos highlighted issues that arise in computational modeling in terms of the mismatch between what can be measured in a medical laboratory and what can be simulated owing to discrepancies in the spatial and temporal scales of the data. Chung asked what types of data are needed to build a DT. Although patient data are sparse, Kalpathy-Cramer stressed the importance of balancing the patient’s best interests with the desire to learn more by collecting only the minimum amount of data necessary for a model to be useful both in terms of input and output. She added that a forward model that generates synthetic data could also be useful.
Chung inquired about the role of a human-in-the-loop to intervene and react to a DT. Glazier pointed out that although the aeronautics industry uses a reactive approach (i.e., if a DT predicts an engine failure, the engine is removed from the aircraft and repaired), the biomedical field would instead benefit from continual interventions to maintain a patient’s healthy state. In the absence of single protocols, this approach would require significant changes in perspectives on medication. He envisioned a future with human-out-of-the-loop regulation of health states but acknowledged challenges in the loss of autonomy and the possibility for catastrophic failure. Self-driving cars provide an important lesson for the future of medical DTs in terms of the value of bootstrapping, he continued. He suggested rolling out DTs even if they do not predict well or far in time, and creating millions of replicas to understand where they fail and how to make improvements. Mechanisms could then be designed to automatically learn from the failures at the population level to improve model parameters and structures.
Chung asked about the pipelines needed for the consistent and accurate parameterization of DT models. Koumoutsakos replied that the first step is to determine the metric for how closely the model represents reality, incorporating uncertainty. The next step is to combine domain-specific knowledge with system-specific data and to leverage AI, ML, and uncertainty quantification techniques. Once the model is developed, he explained, one should be prepared to do iterations and optimizations to the parameters based on the chosen metric.
Given the dynamic nature of DTs, Chung wondered about the time scale of measurement necessary to advance DTs at the organ, tumor, and microenvironment level. Kalpathy-Cramer said that the time scale is determined by the length of time it takes to see a drug response, and she suggested developing strategies to handle multiple time scales. Yankeelov remarked that daily model optimization and prediction is feasible for diseases that require daily imaging; for more systemic therapies, that level of monitoring is far too burdensome for a sick patient. However, he stressed that actionable predictions that might help a person in the near term can still be made without knowing everything about a system. Chung posed a question about the ability to bootstrap limited data sets to build out missing time scales. Glazier responded that in many cases, noninvasive measurements are impossible. Therefore, he emphasized that mixed modeling methods are essential—for example, single-cell models of the origin of resistance and continual models of tumor margins. He also proposed modeling organs-on-a-chip systems to be able to measure in better detail, develop confidence in workflows, and understand failure modes. Because mechanistic simulations can generate what is missing from real-world training data, he asserted that combining mechanistic and data-driven approaches could lead to the development of more reliable models.
Chung inquired about the different challenges that arise when using DTs for diagnosis, prognosis, therapy optimization, and drug discovery. Reflecting on this hierarchy of difficulty, Glazier observed that drug discovery is challenging owing to a lack of treatment models. Furthermore, models act at the molecular level even though the responses of interest occur at the system level, and many levels of interaction exist between the two. Measuring and controlling at the same level of the outcome of interest is impossible in medicine; he noted
![]()
that bulk aggregation could be a useful approach except when two patients who look identical to the model behave differently.
Chung asked about emerging technologies that could be leveraged for DTs as well as significant research gaps that should be addressed. Koumoutsakos added that data obtained from medical applications are often heterogeneous, and a difficult question remains about how to combine them to create DTs. He also emphasized the need to create a common language between the people who take the measurements and those who do the modeling.
DIVERSITY OF MODELS AND DATA IN DIGITAL TWINS
During the third panel, workshop participants heard brief presentations on DTs at the whole-human, multisystem, and population scale from the perspectives of innovative regulatory evaluation and methods development. Much of the conversation centered around the diversity of models and data in DTs.
Aldo Badano, Food and Drug Administration (FDA), indicated that FDA has developed tools to de-risk medical device development and facilitate the evaluation of new technology using least burdensome methods. FDA’s Catalog of Regulatory Science Tools5 includes the Virtual Family6 and the open-source computational pipeline from the first all–in silico trial for breast imaging.
Badano explained that manufacturers are expected to establish the safety and effectiveness of their devices for intended populations—usually via clinical trials. In a digital world, this requires sampling large cohorts of digital humans to enroll digital populations, which can be data-driven and/or knowledge-based. He described knowledge-based models as the most promising for device assessment because they can be incorporated into imaging pipelines that contain device models. He emphasized that digital families have been used for safety determinations in more than 500 FDA submissions, and digital cohorts could be used for in silico device trials. Badano postulated that properly anonymized DT data sets could become rich sources of data for generating digital humans in the future. He added that if DTs are eventually embedded into devices for personalized medicine, the DT model itself would likely need to be incorporated into the regulatory evaluation.
David Miller, Unlearn.AI, commented that despite the skepticism surrounding the use of DTs, they have much scientific potential, especially for the development of prognostic DTs. Prognostic DTs provide a rich set of explanatory data for every participant in a randomized clinical trial: each individual in the trial receives a rich set of predictions (i.e., a multivariate distribution) for all outcomes of interest. He stressed that combining prognostic DTs and real participants enables faster and smaller trials.
Miller explained that this method of applying an ML model to historical data and dropping its predictions into a clinical trial without adding bias (known as PROCOVA™) has been qualified by the European Medicines Agency as suitable for primary analysis of Phase 3 pivotal studies. The procedure has three steps: (1) training and evaluating a prognostic model to predict control outcomes, (2) accounting for the prognostic model while estimating the sample size required for a prospective study, and (3) estimating the treatment effect from a completed study using a linear model while adjusting for the control outcomes predicted by the prognostic model. He emphasized that the context of use for DTs is essential: with zero-trust AI, high correlations provide the greatest value, but weak correlations cause no harm; with minimal-trust AI, low bias (for which a confirming mechanism exists) provides the greatest value; and with high-trust AI, precision medicine could replace clinical judgment.
Discussion
Incorporating questions from workshop participants, Thiele moderated a discussion among both speakers and two experts in bioengineering and personalized oncology, respectively: Todd Coleman, Stanford University, and Heiko Enderling, H. Lee Moffitt Cancer Center. Thiele inquired how simulations, computational models,
__________________
5 The website for FDA’s Catalog of Regulatory Science Tools is https://www.fda.gov/medical-devices/science-and-research-medical-devices/catalog-regulatory-science-tools-help-assess-new-medical-devices, accessed February 27, 2023.
6 The Virtual Family is a package of extremely detailed, anatomically accurate, full-body computer models used in simulations for medical device safety.
![]()
and DTs should be distinguished from one another. Enderling highlighted an overlap in the definitions: DTs need simulation, mechanistic modeling, AI, and ML to answer different types of questions at different scales. He emphasized that synergizing approaches leads to successful outcomes; for example, AI could be used to make mechanistic models more predictive. Coleman suggested that the research community apply its knowledge of anatomy and physiology to develop better generative statistical models and a better understanding of sources of variability. Badano noted that DTs, in silico methods, and AI are distinct concepts that should not be described interchangeably.
Thiele asked about the roles of real-time data, continuous updates, and recalibration for DTs. Coleman stressed that context matters: for a clinical trial related to a neuropsychiatric condition, the DT should be able to predict abrupt changes in behavior and physiology and use that information. He added that the need for continuous updates depends on the richness of the statistics of the population of interest. For applications like PROCOVA™, Miller noted that real-time feedback is not ideal. Enderling and Coleman pointed out that the definition of “real time” (in terms of data collection) is based on the disease of interest. Enderling commented that when studying hormone therapy for prostate cancer, collecting prostate-specific antigen measurements every 2–3 weeks means that the model can be run in between collections, which is sufficient to guide therapy.
Thiele posed a question about the roles of model and parameter standardization for DTs. Enderling replied that calibration and validation are difficult with limited data; he urged the community to identify common objectives and develop rigorous standards to create more trustworthy models. Miller highlighted the value of out-of-sample evaluations, which should match the target population. Badano commented on the need to balance individual validation efforts with population validation efforts.
In response to a question from Thiele about ownership of patient data, Miller stressed that patients should have more control over how their data are shared in clinical trials and used for DTs. Thiele wondered how patients and clinicians could develop trust in DTs. Enderling posited that ensemble models could be used to quantify uncertainty and predictive power and noted that a DT’s predictions should be used as a “communication tool” between the clinician and the patient. Miller championed sharing success stories that start with simple problems: patients and clinicians will not trust DTs until the scientific community has fully endorsed them. Coleman focused on the need to learn from past failures and on ensuring that anatomy and physiology is reflected accurately in algorithms He asserted that in the case of medical applications, knowledge about anatomy and physiology should be reflected accurately in algorithms. Badano explained that simulation enables exploration of patients at the edge of the distribution, which offers another avenue to better understand failure modes. Enderling added that education and transparency within the biomedical community (e.g., developing standardized terminology) are essential before successfully communicating across disciplines and educating clinicians and other stakeholders about the potential of DTs.
Thiele posed a question about the challenges of estimating uncertainties from the population level to the individual level. Badano described a benefit of in silico methods: once the models are running, more data can be integrated, enabling the study of a narrower group without increasing the duration of clinical trials. Coleman referenced opportunities for optimal experimental design and sequential experimental design to improve the performance of digital systems at both the population and individual levels. Miller added that prediction methods at the individual level are in a different regulatory category than those for the population level, which could present barriers to innovation.
Thiele highlighted the interdisciplinarity of DTs and wondered about the potential to develop a culture in which publishing failures is acceptable. Miller doubted that financial incentives would emerge to encourage the private sector to publish failures, but he urged peer reviewers in academia to publish papers with negative findings because they could improve models and advance
![]()
science. Badano suggested modifying the focus from “failures” to “constraints on the results of a model.” Enderling proposed creating a database of negative clinical trials with explanations to help build better predictive models.
CONNECTING DATA AND MODELS ACROSS SCALES
During the fourth panel, workshop participants heard brief presentations on the challenges and opportunities associated with leveraging data across scales and multiscale modeling.
Bissan Al-Lazikani, MD Anderson Cancer Center, defined DTs as computational models that support a clinician’s decision-making about a specific patient at a specific time. The near-term goal for DTs is the development of fit-for-purpose models, with ambitions for the use of multiscale, comprehensive DTs in the future. Current challenges include questions about how to connect DTs across scales, to address vast missing data, and to account for confidence and error.
Liesbet Geris, University of Liège, described the use of intracellular modeling to understand how to shift osteoarthritis patients from a diseased state to a healthy state—a combination of data-driven and mechanistic models enabled a computational framework that could be validated with in vitro experiments. She explained that osteoarthritis is a disease not only of the cells but also of the tissues and the skeleton. A multiscale model of osteoarthritis allows information to be passed from the tissue to the intracellular level, helping to determine whether a cell will stay healthy or remain diseased, and validation occurs at every level of interaction.
Geris also provided an overview of the EDITH7 project, which is building an ecosystem for DTs in health care. The project’s 2022–2024 goal is to build a vision for and roadmap to an integrated multiscale, -time, and -discipline digital representation of the whole body that could eventually enable the comprehensive characterization of the physiological and pathological state in its full heterogeneity and allow patient-specific predictions for the prevention, prediction, screening, diagnosis, and treatment of a disease as well as the evaluation, optimization, selection, and personalization of intervention options. Other project goals include identifying research challenges and enabling infrastructure, developing use cases, and driving large-scale adoption, all of which require advances in technology; user experience; ethical, legal, social, and regulatory frameworks; and sustainability.
Gary An, University of Vermont, noted that some medical DT tasks related to prognosis, diagnosis, and optimization of existing drugs are “scale agnostic.” However, in other cases, such as drug development, understanding how systems cross scales is important. A bottleneck arises in drug development when trying to determine whether the drug that is expected to work will actually work in patients. To reduce the occurrence of failed clinical trials, he suggested enhancing the evaluation process. To evaluate the potential effect of manipulating a particular pathway (proposed for a novel clinical context) as it manifests at a patient level, he asserted that a mechanistic, multiscale model may be needed.
An emphasized that cell-mechanism, multiscale DTs present several challenges, and many questions remain about epistemic uncertainty/incompleteness, high-dimensional parameter spaces, model identifiability/uncertainty, composability/modularity, and useful intermediate applications. He shared a demonstration of a cell-mechanism, multiscale model of acute respiratory distress syndrome that could become a DT if data interfaces to the real world could be established.
Al-Lazikani elaborated that bottlenecks in drug discovery arise owing to the challenges of multidisciplinary and multiscale data integration and multiparameter optimization. To alleviate the issues associated with integrating data from disparate disciplines that span scales, instead of integrating the data points themselves, she suggested integrating how all of the data points interact with each other—essentially establishing edges that can be modeled graphically.8 This approach, which is especially useful when data are sparse, is
__________________
7 The EDITH website is https://www.edith-csa.eu, accessed February 27, 2023.
8 Examples of this approach can be viewed on the canSAR.ai website at https://cansar.ai, accessed February 27, 2023.
![]()
advantageous in that different data are captured in the same logic. It is particularly promising for identify drug-repurposing opportunities and novel therapeutics for cancers such as uveal melanoma. However, a disadvantage emerges with the need to estimate the impact of what is missing; in this case, she asserted that a hybrid approach of data-driven and mechanism-based models would be useful.
Discussion
Incorporating questions from workshop participants, Rebecca Willett, University of Chicago, moderated a discussion with the panelists. In response to a question from Willett about the technical challenges of building multiscale models, Geris pointed out that challenges at the single scale persist when connecting across scales, and finding a way for the scales to communicate is difficult. She stressed that verification, validation, and uncertainty quantification are critical both at the single scale and for the couplings across scales. The EDITH platform aims to establish agreements on the information that should be passed from one scale (or one organ) to another as well as a common language that can be used across modeling platforms and scales. Willett asked about strategies to ensure that a molecular- or cellular-scale model could be plugged in to a multiscale framework with multiple objectives. Al-Lazikani proposed standardizing the interoperability layers instead of standardizing the data or the models themselves. An observed that such issues of composability are an active research gap.
Reflecting on the use of mechanistic models for drug development to target a particular pathway, Willet posed a question about how to build a DT when the pathway is unknown as well as about the value-added of a DT when the pathway is known. An replied that mechanistic computational models are used with perpetually imperfect knowledge to make predictions at a higher level. Because it will never be possible to know everything, he suggested compartmentalizing what is known and validating that knowledge at both a system level and a clinically relevant population level that captures heterogeneity across the population—which is essential for simulating the intervention side of a prospective clinical trial. Al-Lazikani explained that although pathways were useful for drug discovery 20 years ago, now they are insufficient for treating incredibly rare, heterogeneous diseases. A DT of a subpopulation with a rare disease carries the clinical population throughout, so clinical trials can essentially be done in silico to better determine the potential effectiveness of a drug. An described this as an iterative process, with insight into latent functions through the response to perturbations in the clinical setting.
Willett inquired about the role of causal relationships when building models. Geris replied that, in the study of osteoarthritis, information about joint mechanics will interact with the cellular level and the major pathways; thus, both the engineering and systems biology perspectives should work in tandem.
IMPLEMENTATION ISSUES SURROUNDING DATA PRIVACY IN DIGITAL TWINS
Incorporating questions from workshop participants, Qualters moderated the workshop’s final panel discussion among three experts on privacy and ethical considerations for collecting and using data for biomedical DTs: Jodyn Platt, University of Michigan; Lara Mangravite, HI-Bio; and Nathan Price, Thorne HealthTech.
Qualters asked about the benefits of aggregating data for medical DTs and whether additional privacy issues arise beyond those normally associated with health data. Platt highlighted the need to maintain flexibility between knowing what the right data are and having an adaptable data ecosystem, and navigating issues of privacy—protecting individuals from harm should be the primary focus. Mangravite championed the benefits of engaging patients and their care teams more proactively in a conversation about managing care with DTs. She added that collecting data in real time for decision-making creates data governance issues. Price emphasized that combining data threads for DTs could drastically improve personalized medicine; however, increased knowledge about individuals has implications for privacy. He observed that even now, one can push a button on a digital wearable to allow or prevent data
![]()
access across many devices. He reflected on the value of data governance and stressed that all uses of data for DTs should be made known to, understood by, and controlled by the affected individual.
Qualters pointed out that when data are being collected in real time to improve a model and its predictability, bias could creep into those models if significant populations are excluded. Platt urged attention toward issues of equity that arise in terms of individuals’ access to the health care system and to medical technology itself. Patient and community engagement helps to build trust in the benefits of new technology, strengthen accountability, and empower individuals.
Qualters wondered how model uncertainty translates into patient guidance. Price responded that although medical DTs are not yet deployed often in the real world, guidance is based on the data collected at the molecular scale, connected with patient outcome data. Although today this process is human-centric, in the future it might be controlled primarily by algorithms. Therefore, he continued, a relationship with the patient that continues over time is essential to understand prediction accuracy, and guiding the DT feedback loop could help safeguard patients. Qualters asked about non-patient stakeholders (e.g., clinicians, health systems, regulators) who will also have to deal with the uncertainty of DT predictions. Platt encouraged the health system to prepare for the DT experience, as patients expect their doctors to understand and communicate results from the digital space. Mangravite agreed that the opportunity to use DTs for decision-making affects the entire health care system. She stressed that DTs are currently designed to be decision-support tools for humans, who can evaluate and act on the uncertainties.
Qualters invited the panelists to identify research gaps related to privacy and ethical concerns for DTs. Price mentioned the issue of identifiability and supported a focus on the uses of the data. Data-sharing mechanisms are also needed, especially when data are aggregated and models include a person’s complete health history. He stressed that an individual’s DT will never be completely de-identifiable. Mangravite observed research gaps related to data aggregation and interoperability. Once a governance model is developed to control data use, questions will arise about appropriate data and model access.
Qualters inquired about insurability and other economic impacts related to DT predictions. According to Platt, some people fear that their cost of health care will increase or that they will be less insurable based on the DT’s results. Price agreed that insurance companies could benefit by leveraging DTs, and appropriate regulations would be needed. However, he added that if DTs advance preventive medicine, insurance companies that do not adopt them will lose members. He urged workshop participants to focus not only on the challenges of medical DTs but also on the exciting opportunities, such as helping to prevent serious illness in the future.
![]()
DISCLAIMER This Proceedings of a Workshop—in Brief was prepared by Linda Casola as a factual summary of what occurred at the workshop. The statements made are those of the rapporteur or individual workshop participants and do not necessarily represent the views of all workshop participants; the planning committee; or the National Academies of Sciences, Engineering, and Medicine.
COMMITTEE ON FOUNDATIONAL RESEARCH GAPS AND FUTURE DIRECTIONS FOR DIGITAL TWINS Karen E. Willcox (Chair), Oden Institute for Computational Engineering and Sciences, The University of Texas at Austin; Derek Bingham, Simon Fraser University; Caroline Chung, MD Anderson Cancer Center; Julianne Chung, Emory University; Carolina Cruz-Neira, University of Central Florida; Conrad J. Grant, Johns Hopkins University Applied Physics Laboratory; James L. Kinter III, George Mason University; Ruby Leung, Pacific Northwest National Laboratory; Parviz Moin, Stanford University; Lucila Ohno-Machado, Yale University; Colin J. Parris, General Electric; Irene Qualters, Los Alamos National Laboratory; Ines Thiele, National University of Ireland, Galway; Conrad Tucker, Carnegie Mellon University; Rebecca Willett, University of Chicago; and Xinyue Ye, Texas A&M University-College Station. * Italic indicates workshop planning committee member.
REVIEWERS To ensure that it meets institutional standards for quality and objectivity, this Proceedings of a Workshop—in Brief was reviewed by Ahmet Erdemir, Computational Biomodeling Core, Cleveland Clinic; Irene Qualters, Los Alamos National Laboratory; and Adrian Wolfberg, National Academies of Sciences, Engineering, and Medicine. Katiria Ortiz, National Academies of Sciences, Engineering, and Medicine, served as the review coordinator.
STAFF Samantha Koretsky, Workshop Director, Board on Mathematical Sciences and Analytics (BMSA); Brittany Segundo, Study Director, BMSA; Kavita Berger, Director, Board on Life Sciences; Beth Cady, Senior Program Officer, National Academy of Engineering; Jon Eisenberg, Director, Computer Science and Telecommunications Board (CSTB); Tho Nguyen, Senior Program Officer, CSTB; Patricia Razafindrambinina, Associate Program Officer, Board on Atmospheric Sciences and Climate; Michelle Schwalbe, Director, National Materials and Manufacturing Board (NMMB) and Board on Mathematical Sciences and Analytics; and Erik B. Svedberg, Scholar, NMMB.
SPONSORS This project was supported by Contract FA9550-22-1-0535 with the Department of Defense (Air Force Office of Scientific Research and Defense Advanced Research Projects Agency), Award Number DE-SC0022878 with the Department of Energy, Award HHSN263201800029I with the National Institutes of Health, and Award AWD-001543 with the National Science Foundation.
This material is based on work supported by the U.S. Department of Energy, Office of Science, Office of Advanced Scientific Computing Research, and Office of Biological and Environmental Research.
This project has been funded in part with federal funds from the National Cancer Institute, National Institute of Biomedical Imaging and Bioengineering, National Library of Medicine, and Office of Data Science Strategy from the National Institutes of Health, Department of Health and Human Services.
Any opinions, findings, conclusions, or recommendations expressed do not necessarily reflect the views of the National Science Foundation.
This proceedings was prepared as an account of work sponsored by an agency of the United States Government. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States Government or any agency thereof. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof.
SUGGESTED CITATION National Academies of Sciences, Engineering, and Medicine. 2023. Opportunities and Challenges for Digital Twins in Biomedical Research: Proceedings of a Workshop—in Brief. Washington, DC: The National Academies Press. https://doi.org/10.17226/26922.














