To assess the functional consequences of genetic variants, human models of disease are essential, particularly when studying a combination of rare and commengenetic variants, said Dimitri Krainc. Not only is the effect size small for many of these variants, but effects may also vary across different genetic backgrounds, he said. Krainc gave an example of the need for human-based studies from the Parkinson’s disease (PD) research conducted in his own lab. The cardinal symptoms of PD—bradykinesia, tremor, and rigidity—result from the degeneration of dopamine neurons in the substantia nigra. Research in Krainc’s lab has shown that dopaminergic neurons from patients with PD accumulate oxidized dopamine, which leads to, lysosomal dysfunction, and α-synuclein accumulation, yet nigral dopaminergeic neurons in mouse models of PD show no accumulation of oxidized dopamine and appear resistant to degeneration in genetic models of disease, said Krainc (Burbulla et al., 2017). In other words, he said, “It’s a fundamentally different physiology and pathology in human and mouse dopamine neurons.” Moreover, genetic studies of PD suggest that many genes implicated in the disease cluster around mitochondrial, lysosomal, and synaptic function, yet mouse models would not be useful in studying these pathways.
Krainc said his lab now has completely stopped using mouse models of PD for studies of vulnerable dopaminergic neurons and is focused on neurons from patients. While these cell models are sufficient to study cell-autonomous dysfunction of dopamine neurons, as well as interactions with glia, they do not allow for studies of more complex cell–cell interactions and brain circuits that play important role in the pathogenesis of PD.
Thus, other models are needed, including novel animal models, nonhuman primates, organoids, he said.
MODELING IN FROGS
To understand how to treat psychiatric disorders, identifying the relevant brain regions, cell types, timepoints, and functions that are affected is essential, said Helen Willsey, an assistant professor at the Weill Institute for Neurosciences at the University of California, San Francisco (UCSF). Foundational research based on human genetics is needed to pinpoint these characteristics, she said.
For example, in autism spectrum disorder, genetic studies have implicated common genes with small effect sizes as well as many rare variants with large effect sizes, said Willsey. Each one of these genes represents an opportunity to answer questions regarding relevant cell types, brain regions, and functions as a precursor to rational drug design. The complication is that many genes contribute and each one is highly pleiotropic, meaning it plays multiple roles in development, she said. What is needed is a model that enables investigation of multiple genes in parallel during brain development. Willsey argued that phenotypes common to multiple different genes are more likely to be relevant to a disorder’s pathobiology.
Willsey has developed a high-throughput genetic model system in the frog Xenopus tropicalis, which like humans has a diploid genome. One mating pair of this frog can produce, in just one afternoon, more than 4,000 embryos that develop outside of the mother. The embryos can be exposed to drugs simply by adding the drug to the water, making it a valuable model for drug discovery. Willsey and colleagues have made half-mutant animals by taking embryos at the two-cell stage and injecting gene editing reagents and dye into one cell. The resulting tadpoles have one half of the brain mutated with the gene of interest and labeled with dye, enabling comparison within the animal of the consequences of the mutation on brain development. Willsey said it takes only 5 days to get free-swimming, behaving animals, making this model both efficient and cost effective. Moreover, Xenopus makes many different cell types, allowing investigators to look at and compare mutated versus wild-type proliferative progenitor cells, excitatory and inhibitory neurons, astrocytes, glia, and microglia within the same animal. Willsey and colleagues have also developed different three-dimensional imaging methodologies to examine these different cell types in the Xenopus brain (see Figure 5-1).
Using the Xenopus model, Willsey and colleagues have shown that all 10 of the most statistically associated risk genes for autism cause changes in brain size and particularly in forebrain neurogenesis when neurons are made. This study suggested that neurogenesis may have some relevance to
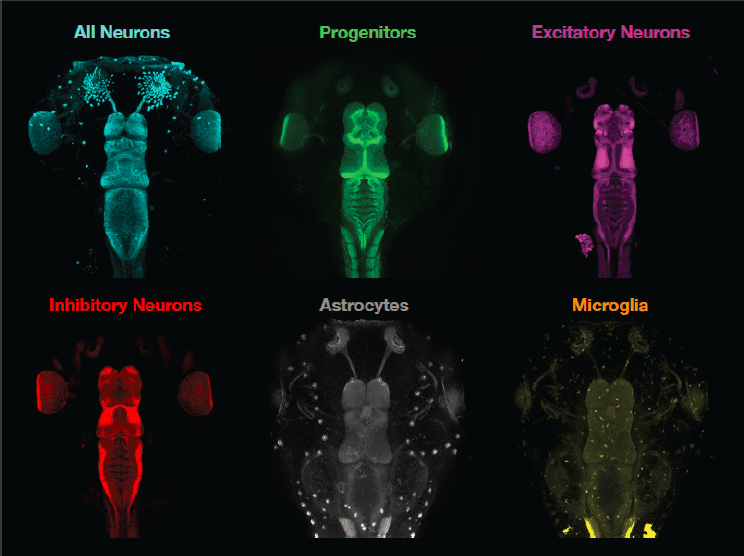
SOURCE: Presented by Helen Willsey, October 6, 2021.
autism pathology, said Willsey. They also showed they could rescue this phenotype with estrogen, which she suggested could explain at least in part the well-known male predominance in autism.
To validate the Xenopus findings in more human-derived settings, Willsey and colleagues showed that the highest level of network connectivity of autism risk genes was in the subventricular zone, a very neurogenic layer of the cortex. In induced pluripotent stem cells– (iPSC-) derived neural progenitor cells, they recapitulated the proliferation defect of four autism risk genes; in addition, in human cortical organoids, they showed that estrogen rescued the inhibition of the autism risk gene DYRK1A, which contributes to decreased forebrain size in Xenopus.
Willsey added that the Xenopus model not only enables the generation of foundational knowledge and hypotheses that can be followed up in human models, but also allows investigation of the effects of genes in other organ systems. For example, individuals with autism often have gastrointestinal dysfunction and congenital heart disease, both of which can be
studied in frogs. Moreover, said Willsey, because frogs behave in interesting and complex ways, they facilitate investigation of how fundamental changes in molecular mechanisms translate into circuit-level functional changes and behavior.
MODELING IN NON-HUMAN PRIMATES
As Willsey demonstrated, some aspects of deeply conserved biology are relevant to human disorders that can be studied in evolutionarily distant animal models. However, when there is a lack of deep conservation, or where in vitro models may be inadequate, non-human primate models are valuable, said Fenna Krienen, postdoctoral fellow in Steve McCarroll’s lab at Harvard Medical School and The Broad Institute. Krienen uses single-cell nucleus sequencing from postmortem brain tissue as well as tissue from mice, ferrets, macaques, and marmosets.
Inhibitory interneurons are one major cell type Krienen and colleagues have studied. These cells are conserved across mammals, with the same strong and selective markers and genes, she said; however, there are many qualitative and quantitative changes in gene expression. These differences reflect the evolution of the genetic programming of inhibitory interneurons, as illustrated in Figure 5-2 for the gene NTNG-1 (netrin G1), a gene that has been linked to syndromic autism.
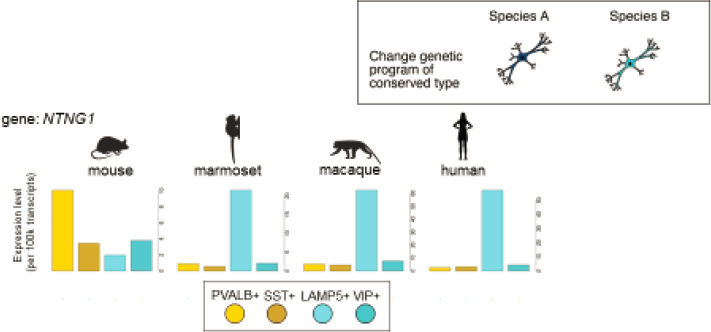
NOTE: LAMP5 = Lysosome-associated membrane glycoprotein 5; NTNG-1 = netrin G1; PVALB = parvalbumin; SST = somatostatin; VIP = vasoactive intestinal peptide.
SOURCE: Presented by Fenna Krienen, October 6, 2021.
Krienen and colleagues have also compared gene expression between pairs of species, showing, for example, that expression of another autism candidate risk gene, NRG1 (neuregulin 1) in human interneurons, can be predicted by its level of expression in non-human primates, but not in mice or ferrets. These examples illustrate that evolutionary proximity constrains the expression even of genes that are thought to be essential for normal neuronal function, said Krienen.
Another way brain structures may evolve is via a novel cell type, said Krienen. Again focusing on interneurons and studying the striatum, which is thought to be deeply conserved, she and her team identified a previously unknown interneuron expressing the neuropeptide TAC3 (Krienen et al., 2020). TAC3-positive interneurons are not seen in mouse brains, she said, and the neuropeptides and transcription factors they express are not seen in other interneuron subtypes in mice or marmosets. Moreover, TAC3-positive neurons comprise as much as 30 percent of striatal neurons in primates. She added that the pattern of gene expression and epigenetic expression in these neurons are categorically different from other interneuron types, suggesting that it may have evolved novel regulatory features early in development. However, she said, new tools are needed to further characterize it.
Krienen said the kind of information gained from single-cell sequencing approaches enables scientists to make rational, informed choices about the models to use in studying human brain biology. These approaches also have translational use, she said. They enable nomination of candidate enhancers and, using high-throughput and barcoded approaches, facilitate the development and packaging of candidate enhancers into pools of adenoassociated viruses, she said. Single-cell sequencing also provides readouts for demonstrating cell-type specificity and enables the screening of potentially dozens or hundreds of enhancers in a single animal, thus minimizing costs and animal use, said Krienen.
HUMAN BRAIN ORGANOIDS
Integrating and leveraging genetic information about neurological and psychiatric illnesses is essential to understanding disease mechanisms, said Paola Arlotta, chair and Golub Family Professor of Harvard’s Department of Stem Cell and Regenerative Biology. To achieve this, she advocated for an expanded range of experimental models of the brain to enable hypothesis testing and mechanistic discovery. Models of the human brain are especially valuable, she said, not only because the human brain differs substantially from that of rodents and other experimental organisms, but especially because studying these genetically complex disorders in which many risk variants are spread across the genome requires models that represent the human genome, said Arlotta.
Arlotta and colleagues have generated from pluripotent stem cells (iPSCs and ESCs) three-dimensional, multicellular, human brain “organoids,” which enable them to conduct a spectrum of studies aimed at understanding aspects of human brain development, mechanisms of neuropathology, and gene effects on the functionality of the human brain. Eventually, she said, this platform may also enable chemical screening. Early organoids created in the Arlotta lab were able to develop over extended periods of time, generating a large diversity of cell types and networks of neurons with interesting electrophysiological properties, said Arlotta (Quadrato et al., 2017). Initially, organoids were scalable systems, but not very reproducible, she said. Further work in her lab eventually yielded organoids with the diversity of cell types present in the developing cerebral cortex that could also be developed in a consistent and very reproducible way, said Arlotta (Velasco et al., 2019). The Arlotta lab has since shown how this model informs understanding of human disease (Paulsen et al., 2020).
For example, Arlotta and colleagues generated organoids carrying three highly penetrant risk mutations in autism spectrum disorder. This work, published as a preprint, demonstrates how they used the organoids to identify affected cell types and biological process, determine when cells were affected during development, examine whether and how these three genetic mutations converged to produce a phenotype, and assess whether and how the genetic background in which the mutations acted influenced the disease phenotype.1 What they found was that through largely different molecular mechanisms, all three of these genes converge on asynchronous development and maturations of two neuronal populations of the cerebral cortex, GABAergic neurons and deep layer projection neurons, said Arlotta. They also showed that the genetic background of the donor stem cell line affected the phenotype, indicating that individual genomes can modulate the effect of even highly penetrant risk genes. Arlotta concluded that these organoids serve as a reductionist and reproducible experimental system of the developing human brain, enabling the identification of convergent mechanisms and affected developmental processes. The next step, she said, will be to use the organoids to examine how these early developmental abnormalities affect cortical function.
CRISPR-BASED FUNCTIONAL GENOMICS IN IPSC MODELS
To address the fundamental question of how genes contribute to disease requires consideration of variants in the genome, the impact of background and ancestry, genetic modifiers of disease, and differential expression of genes
___________________
1 Made available in preprint format. To learn more, see https://www.biorxiv.org/content/10.1101/2020.11.10.376509v1 (accessed December 10, 2021).
that may drive disease-related functional outcomes such as selective vulnerability of different cell types, said Martin Kampmann, associate professor of biochemistry and biophysics at the UCSF Institute for Neurogenerative Diseases and an investigator at the Chan Zuckerberg Biohub. Moreover, he said, understanding these disease mechanisms can enable stratification of patients in clinical studies as well as identification of new therapeutic targets.
Kampmann and colleagues have developed scalable, high-throughput, and quantitative functional genomics technologies that perturb genes using a variation of CRISPR-Cas92 technology. Their approach controls the expression of genes targeting transcriptional activators (CRISPRa) or transcriptional repressors (CRISPRi), said Kampmann (Gilbert et al., 2014). Using iPSCs, CRISPRi and CRISPRa have been used to model diseases in neurons, microglia, and astrocytes, he said. Kampmann described two broad classes of experiments that use these tools: (1) high-throughput modeling of genetic variants, and (2) genome-wide modifier screens of disease phenotypes (Kampmann, 2020).
For example, Kampmann’s lab has been using CRISPRi to screen the entire genome for modifiers of tau aggregation, one of the hallmarks of Alzheimer’s disease (AD) and other tauopathies. In collaboration with Li Gan at Weill Cornell Medical College, they have turned iPSCs carrying different tau mutations into neurons. They then use CRISPRi to target thousands of protein-coding genes and can then screen the neurons for tau aggregation. Next-generation sequencing then allows them to identify genes that modulate tau aggregation. Kampmann said this approach has allowed them to find modifiers of both tau production and tau oligomerization that could turn out to be therapeutic targets.
Microglia have also been implicated as potential drivers of neurodegenerative diseases such as AD, said Kampmann. To better understand the role of distinct microglia cell states in AD, Kampmann and colleagues have established a screening platform combining iPSCs with CRISPRi/a machinery to identify which genes push microglia toward or away from cell states linked to different disease phenotypes (Dräger et al., 2021).
MACHINE LEARNING
Although genetics is known to suggest causal mechanisms through the effects of genes on human phenotypes, the many variants implicated
___________________
2 Clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9 (CRISPR-Cas9) is a widely used genome editing technology that is fast, inexpensive, accurate, and efficient. To learn more about CRISPR-Cas9, see https://www.broadinstitute.org/what-broad/areas-focus/project-spotlight/questions-and-answers-about-crispr (accessed December 10, 2021).
in complex traits along with their small effect sizes present challenges to investigators trying to determine which genes actually have a meaningful effect on human clinical outcomes, said Daphne Koller, CEO and founder of Insitro, a machine learning-enabled drug discovery company. Insitro uses machine learning to create an intermediate point between genetics and clinical outcomes by using high-content biological data—both clinical data and functional data collected from cells derived from human genetics—as a bridge, she said.
Modern-day machine learning has progressed well beyond early approaches where investigators defined what features to look at in a dataset, said Koller. The problem with that approach, she said, is that people are not very good at defining predictive features, especially when analyzing biological data. Now, said Koller, machine learning starts with raw data and via a process called end-to-end training, figures out for itself an increasingly complex hierarchy of features built on other features, eventually yielding a prediction. “The machine is able to identify features that are much more subtle and salient than a person would ever be able to do,” said Koller.
The machine also uses high-dimensional features to define low-dimensional “manifolds” in which the data are embedded, said Koller. By capturing similarities that may not be obvious via embedding in a new manifold, the machine effectively can uncover new structures and insights in biological data, she said. For example, Insitro has been working with Gilead to try to identify genetic drivers of fibrotic progression in nonalcoholic steatohepatitis (NASH) using high-content histological data from patients at the beginning and end of a clinical trial. Koller said the machine learning model was able to extract data on the patient state that is much more granular than what a clinician is able to ascertain, and thus was able to identify patterns that were otherwise invisible. “We were able to identify, for the first time, two novel genome-wide significant variants of fibrotic progression in NASH that have very compelling biology and are eminently prosecutable as drug targets,” said Koller.
The insights derived from human data are then used to build in vitro cellular systems that recapitulate the complex polygenic architecture of human disease, said Koller. Machine learning then takes the data obtained from these cellular systems to build phenotypic manifolds that inform predictive disease models, she said (see Figure 5-3).
The phenotypic manifold may also be used to identify interventions that might modulate a disease, said Koller. For example, she said they have created an isogenic iPSC-derived model of tuberous sclerosis complex (TSC), a developmental neurological disorder that causes refractory epilepsy and is caused by loss-of-function mutations in TSC1 or TSC2 genes. Machine learning has enabled identification of a clear but previ-
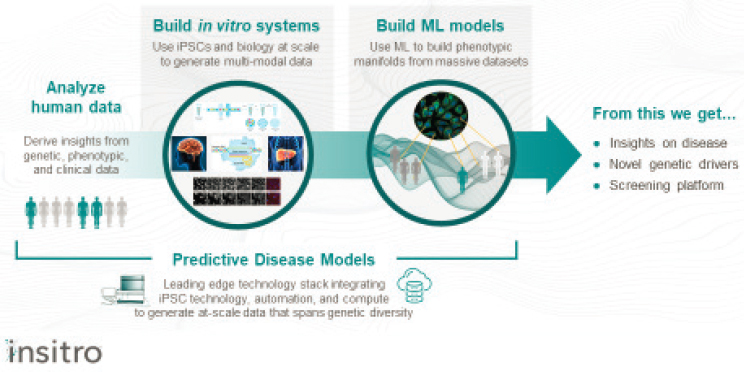
SOURCE: Presented by Daphne Koller, October 6, 2021.
ously unrecognized phenotype that differentiates cases and controls, and has contributed to the validation of a gene that appears to be a therapeutic target for the disease, she said. She added that they are also using the platform to uncover different pathways that cluster together in phenotypic space for amyotrophic lateral sclerosis (ALS), a much more complex polygenic disease. “The idea is to use this approach to identify a conserved pathophysiology across heterogeneous genetic causes where we can discern coherent subsets of patients that are likely to respond to a single treatment, and then screen for high-impact genetic modifiers of disease in those subgroups,” she said.
ARTIFICIAL INTELLIGENCE
Leveraging the huge amount of human genetic data generated over the past decade, integrating it with other types of human data, and extending it to larger diverse patient populations will increase the probability of clinical success, said Alice Zhang, CEO and co-founder of Verge Genomics. To accomplish this, Verge has collected a large proprietary human tissue database of more than 6,500 individual patient brain samples with clinical metadata, and has genotyped and RNA sequenced the full transcriptome for each sample, said Zhang. By probing this data with well-defined human
genetic drivers where highly penetrant mutations are known to cause disease, they have found disease-causing pathways, revealed the functional consequences of mutations, identified upstream intervention points that can be targeted to normalize genetic pathways, and validated these targets in human-derived iPSC neurons. Using these iPSCs, they are able to determine if drug candidates have disease-modifying potential by restoring neuronal function or slowing neuronal death, said Zhang.
For example, Verge scientists and their collaborators analyzed spinal cord tissue from 983 patients to identify a gene network enriched for genes related to endolysosomal function that was highly associated with C9orf72, the most common genetic cause of ALS, said Zhang. They showed that loss-of-function mutations in C9orf72 lead to increased aggregation of toxic proteins and accumulation of glutamate receptors at the cell surface, which together trigger neurodegeneration (Shi et al., 2018). The scientists further showed that the kinase inhibitor PIKFYVE is a key regulator of the pathway and a potential therapeutic target, and that an inhibitor of PIKFYVE restored neuronal function and rescued survival in ALS patient neurons, said Zhang.
Believing that endolysosomal dysfunction might underlie ALS not only in patients with C9orf72 mutations but in other forms of ALS as well, the researchers tested PIKFYVE inhibitors in multiple genetic models of the disease, said Zhang. Not only did these compounds demonstrate strong and reproducible efficacy across multiple models, but in head-to-head comparisons also outperformed previously developed ALS drugs in the clinic as well as the only two drugs currently approved for ALS, riluzole and edaravone, she added. They also showed that these compounds down-regulated the C9 disease signature in patients with sporadic ALS.
Although the Verge drug discovery platform is what Zhang calls an “all-in-human platform,” in that it combines data from human genetic studies, human tissue, human-derived neuronal cells, and human clinical studies, she said they also employ mouse models to develop a panel of biomarkers that can be translated to early clinical studies to demonstrate target engagement and effects on the downstream biology. For their lead candidate PIKFYVE inhibitor, these biomarkers have demonstrated full target engagement and a reduction in markers of lysosomal dysfunction and neurodegeneration, she said. She added that Verge plans to file an investigational new drug application in 2022 and enter clinical trials shortly afterward.
INTEGRATING DATA FROM A SPECTRUM OF MODELS TO GAIN MAXIMUM INSIGHT
New models such as brain organoids that incorporate different cell types will enable modeling of some disease processes, but other processes will require animal models, said Kampmann. Arlotta added that there will always be a limit to what a single model can do. Human stem cell derived models such as organoids are the only ones that actually model the human genome, she said, but there is a limit to the precise type of neural network analysis that can be achieved with them. Arlotta suggested that chimeric systems may be needed, whereby cells produced in vitro are transplanted in vivo to be able to read higher order properties in the brain. “As a field, we have to move from developing multiple models to assuring that the models are actually reproducing endogenous biology, and to making sure there is reproducibility,” she said. Next-generation organoids will likely contain a better representation of cell types and structures, including vasculature, to model higher-order, more complex events, she said, “I think it’s just a matter of time.”
Steven Hyman agreed about the need to match questions with models, noting that for many questions, a living brain, and thus animals are needed, but that many human disease-relevant processes cannot be recreated in animals, even in nonhuman primates given the need for human genes, including human genetic backgrounds. For example, 90 percent of GWAS hits are noncoding and thus reside in the least well-conserved parts of the genome. Yet, some important features of disease can be recapitulated in particular animal or cellular models, indicating the need for principled integration, he said. The question in modeling any mechanism Hyman asked is, how close is enough to be useful? What kind of validation is needed to move from a model of mechanisms toward a therapeutic? For cellular models, how well can they reflect the relevant maturation state of the brain?
Indeed, said Kampmann, a big challenge in iPSC modeling is getting cell types that are mature enough to reflect particular biological processes. However, he noted that because genetic changes have molecular consequences across the life of the cell, understanding even the earliest changes that are disease relevant could yield hypotheses on how to intervene in early stages of disease. When the first successful example of translating iPSCs to an effective therapeutic in humans is achieved, he said, we will have validated that this approach works. “We are still waiting for the first breakthrough to know what cellular models are valid,” he said.
Koller added another dimension to Kampmann’s comments. The goal of any model system is to predict the human in vivo clinical outcome, yet having a model that faithfully replicates all of the biology of the human organism may not be necessary, said Koller. Machine learning aims to
capture phenomena observed in the in vitro systems, looking at the effects of different interventions on cells from different genetic backgrounds as a means of predicting human clinical outcome, she said. So even if the in vitro system does not capture the biology precisely, it may predict downstream cellular processes that may enable prediction of important disease characteristics such as age of onset or disease severity, she said.
“To me, human genetics is a form of ground truth, so if we can look at the effect of human genetics on a cellular phenotype and use what we see in that cellular phenotype to predict a human clinical outcome that we care about, that is a ground truth test for whether our cellular system is predictive of the human clinical outcome,” said Koller. She added that genetics may also be useful to understand which environmental exposures and other stressors that may be disease relevant. The importance to disease of environmental exposure can be powerful at unmasking molecular phenotypes, added Kampmann.
Willsey added that being able to study multiple genes associated with a disorder in parallel over time and space in different model systems may not only bridge the gaps between model systems, but also help identify a path forward to understand pleiotropy, where genes regulate different functions depending on the cell type and time during development.
Zhang, however, said that although preclinical models may be useful to show proof of mechanism and proof of pharmacology, they are not able to demonstrate proof of clinical efficacy. Genetics can provide guidance regarding causal mechanisms, but linking that mechanism to efficacy can only be done in humans, she said. The goal, she continued, is to get to humans as quickly as possibly by de-risking everything that prevents hypothesis testing in humans.
The field has moved toward a biomarker-driven approach in early clinical studies rather than looking at behavioral readouts, said Zhang. She agreed with Kampmann that proximal observations are more likely to translate than distal ones. For example, she said, if a repeat expansion in a protein leads to toxic aggregates, a drug that removes those aggregates is more likely to translate to the clinic.
Whether the field is actually ready to move forward without behavioral endpoints is another question, said Amir Tamiz, associate director at the National Institute of Neurological Disorders and Stroke and director of the Division of Translational Research. Funders control the limiting factor in this regard, according to Zhang. If investors still want to see behavior and survival, we have to collect that information, she said. Hyman countered that behavior may be a very high-level readout of circuit activity after many levels of integration. The problem, he said, is with how behavioral readouts are interpreted. Willsey agreed, but noted that if a treatment were able to rescue a cellular phenotype in an animal model as well as something
like circadian rhythm dysfunction in humans, the animal model might be useful to study a mechanism for the behavioral dysfunction.
Integrating data from multiple datasets in order to understand pathway biology can also point to prognostic and diagnostic biomarkers that can be used for patient stratification and assessment of treatment response, said Zhang. Koller agreed, noting an example from her work with polygenic liver disease where a subset of patients with a particular genetic variant, who exhibit characteristic morphological and histopathological features, may be separately treatable using a precision therapeutic.
Moving away from highly penetrant genetic variants that affect only a small subset of patients and into a larger patient population that has a disruption in the same pathway is, what Koller called, “the holy grail” in the field. That is where she thinks high-content data from diverse patient populations may reveal convergence at the genetic level. “It’s a gamble,” she said. “We haven’t demonstrated it conclusively, and certainly not in neuroscience.” However, she cited the “poster child of genetic diseases outside of oncology,” PCSK9 mutations, which were discovered in a small number of families affected by autosomal dominant hypercholesterolemia, but led to the development of PCSK9 inhibitors for the treatment of lipid disorders in a wide range of patients (Jaworski et al., 2017). Hyman, however, noted that while PCSK9 was a rare variant that led to a therapeutic, there will be other cases where rare penetrant variants will have muted or different effects within the general population. He pointed to what he called “the tragic example of overfocus on familial Alzheimer’s disease,” where variants in the amyloid precursor protein (APP) and presenilin genes responsible for cleaving APP led the field to focus drug development almost exclusively on targeting amyloid beta (Aβ).
Zhang maintained that another lesson was learned from the overfocus on amyloid. “We didn’t understand the context and the pathway within which Aβ is operating,” she said. “And without a systems understanding of what’s happening at a pathway level, it will be hard to sort those out.” Koller added that there is still an incomplete understanding of how Aβ therapeutics function and that they have not demonstrated significant success even in familial early-onset patients. “So it may be that the problem isn’t that our genetics was wrong, but that our therapy is not actually tackling the right aspect of the process,” she said. Again, said Koller, this is where high-content phenotypes may be useful.














