2
A Multiplicity of Descriptors in Genetics and Genomics Research
INTRODUCTION
Human populations can be described according to countless characteristics: urban versus rural, for example, or smokers versus nonsmokers. With the use of descriptors such as ethnicity, ancestry, or race, however, the focus is on what can be called descent-based or descent-associated groupings: populations whose members are thought to share some characteristic that derives from their common origin.1 Many kinds of classifications fit this bill; clan, caste, and tribe are other examples. In short, human beings across the globe have devised a family of descent-associated categorization systems (Wimmer, 2013).
As a result, genetics researchers who deem it necessary to incorporate some kind of population descriptor in their work face a choice about which descriptor(s) to use and why. The word choice is crucial here because the inclusion of population descriptors in genomics must be a deliberate decision, not a reflex or an afterthought. Groupings of human beings are ultimately cultural constructs that are closely tied to historical and prevailing political, economic, and other conditions, and their use has the potential to have a deep, direct effect on the user’s ways of seeing and understanding the social world, specifically as it relates to research participants.
Just as importantly, the many—at times conflicting—social meanings that have been attached to descent-associated groups (for example, prior use of inferior or superior, pure or polluted) have added erroneous con-
___________________
1 See Hollinger (1998) on “communities of descent,” Chandra (2012) on “descent-based attributes,” and Morning and Maneri (2022) on “concepts of descent-based difference.”
notations to them that impede scientific understandings of human biological variation. Certainly, the parsing of over 8 billion human beings into four color-coded racial categories rooted in ancient Greek humoral theory amounts to an exceedingly blunt tool for describing the human species (Hannaford, 1996). Moreover, the cultural sensitivity of human classification schemes has led to the use of some population descriptors—such as ethnicity and ancestry—as euphemisms for others, notably race, which in turn blurs the conceptual boundaries between them and makes it hard to know exactly what any given descent-associated term is intended to represent.
Accordingly, this chapter presents multiple descent-associated population descriptors and offers a definition of each that delineates it from the others (see Box 2-1 for brief definitions of relevant terms). As members of the broad family of descent-associated concepts, each descriptor has similarities to one or more of the others, yet each also revolves around a somewhat distinct understanding of human difference and thus offers researchers a specific tool that is more tailored for some uses than for others. The committee’s objective is, then, to briefly describe the population descriptors that emerged from the study as most relevant to its charge and define them in such a way that scientists and others clearly understand what they can capture—or not—in genetic analyses. Note that these descent-associated measures have a long history of use in genetics, but the rationale for using them has rarely been explicit.
This chapter begins by discussing the concept of ancestry, as it is central to all descent-associated classification and is closely tied to notions of inheritance. Geography is considered next, as it is an equally integral—if less often explicitly recognized—element in the conceptualization of descent-associated groupings. After these two fundamental building blocks, the chapter delves into three population descriptors that are prominent in the United States and which all build on ideas of ancestry and geography: ethnicity, indigeneity, and race. Of note, some of these descriptors have multiple dimensions themselves: geography can refer to place of birth, residence, or ancestral origin; ethnicity might be measured by language, dialect, or religion, for example. Hence it is important to think clearly about the myriad possibilities for population descriptors and make a reasoned, deliberate choice when conducting genomics research.
In addition, this chapter considers the conceptualization and inclusion of environmental variables in human genetics and genomics research. As noted in Chapter 1, most phenotypes are the result of interplay between genetic and environmental factors. Human beings might be categorized, for example, according to their exposure to environmental pollutants and thus their risk for asthma. In other words, environmental factors may lead to the identification of different populations, such as high risk and low risk, that
researchers consider meaningful for their analyses. Although environmental factors are distinct from genetic ones, their joint and interactive effects on phenotypes compel us to consider them simultaneously. The final section of this chapter explores the inclusion of environmental variables in genetics research, which could be used independently or along with other descriptors.
A RANGE OF DESCENT-ASSOCIATED POPULATION DESCRIPTORS
Ancestry
Each person has a family tree, a set of biological ancestors consisting of their parents, grandparents, great-grandparents, and so forth. These recent genealogical ancestors are often of interest for their sociocultural and biological heritage. Going back in time, their number rapidly balloons and, because there have been only so many people on the planet, the genealogical ancestors of one person quickly begin to overlap with those of another (Figure 2-1). By several thousands of years ago, all modern humans likely share all their genealogical ancestors (Rohde et al., 2004). In other words, modern humans are all embedded within the same enormous family tree, with some members more closely related than others (Figure 2-1).
For a given individual, most of their remote genealogical ancestors did not transmit DNA to them. In fact, only a tiny subset of a person’s genealogical ancestors many generations back are also their genetic ancestors contributing DNA. At a given position in the genome, a person carries two copies of DNA, inherited from only two of their many genealogical ancestors: for example, they might carry their paternal grandfather and their maternal grandmother’s DNA. Which genealogical ancestors contributed DNA changes along the genome. These changes result from two processes: the fact that only half of the genetic material of a parent is transmitted to an offspring and because the transmitted material is itself the result of recombining the parental DNA. Each person’s genome is thus a mosaic
of DNA segments inherited from a small subset of genealogical ancestors (Donnelly, 1983).
As a result, a person’s genetic ancestry can be thought of in terms of lines extending upwards from one ancestor to another in the family tree (see Figure 2-1). From one position in the genome to another, these lines through the family tree will differ, owing to recombination, traversing a distinct succession of genealogical ancestors. When comparing the genetic ancestry of two individuals, the lines of inheritance will coalesce quickly at some positions and remain distinct for longer times at others (Hudson, 1990; Wohns et al., 2022). Segments of the genome inherited by two or more people from the same relatively recent ancestor are referred to as “identical-by-descent” (Thompson, 2013).
Individuals in greater geographic proximity will tend to share more lines of ancestry through their family tree (Coop Lab, 2017b). Eventually, all modern humans will share a genetic common ancestor at any given position in the genome. For a set of individuals, the genetic ancestry lines that trace to the common ancestor at every position in the genome comprise the ancestral recombination graph (Hudson, 1990; Schaefer et al., 2021), which can also be understood as a sequence of inheritance paths, called gene “trees” for each segment of the genome (Kelleher et al., 2016; Wiuf and Hein, 1999) (see Figure 2-1 inset). The genetic ancestries of all humans today, no matter how or where they are identified, are embedded within this giant graph.
As this brief summary clarifies, any description of the genetic ancestry of an individual entails a decision about the relevant time depth at which to describe it. For example, given the repeated mixing and long-range migrations that have characterized all human evolution (Reich, 2018), individuals living in close proximity in Europe today, who might be characterized as of some particular regional ancestry or of “European ancestry,” trace much of their genetic ancestry also to both central Asia and the Middle East only 8,000 years ago (Haak et al., 2015), and farther back in the past their genetic ancestors lived in Africa (Wohns et al., 2022). Therefore, referring to people with recent genealogical or genetic ancestors in Europe as “white” or of “European ancestry” and people with recent genealogical ancestors in Africa and often Europe as “black” or of “African ancestry” is incomplete, incorrect, and misleading. Similarly, any geographic or ethnic labeling requires that decisions be made about the appropriate level of resolution necessary for a study. In many current studies, individuals have been defined in terms of continental ancestry on a timeframe before the past 600 years, e.g., before the onset of European global colonialism and its associated voluntary and forced population movements, admixture, and the large-scale institution of enslavement.
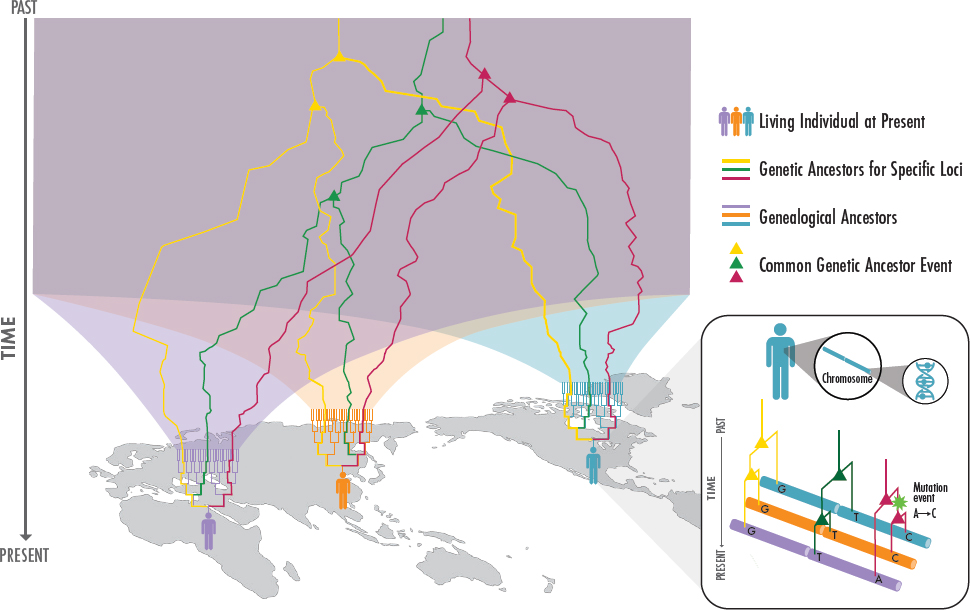
These difficulties are compounded when ascribing population descriptors to groups, as boundaries need to be imposed to divide into bins people who share both genealogical and genetic ancestors. A common approach is to define genetic ancestry groups by first forming clusters of individuals of sufficient genetic similarity based on some genetic measure (e.g., using ancestry proportion inference, see Rosenberg et al., 2002, or principal component analysis, see Reich et al., 2008). These genetic groupings are often then labeled by reference to a nongenetic property that is common in the group, such as a particular geographical origin or ethnicity. Much of the challenge of population descriptors arises from this practice of associating nongenetic terms to clusters of genetically similar individuals and doing so inconsistently within and between studies.
The population descriptors by which individuals are grouped sometimes derive from conceptual constructs used in statistical models. For instance, many statistical methods to infer clusters or groups posit the existence of discrete populations with distinct patterns of genetic variation from which individuals derive their genetic ancestry (Pritchard et al., 2000). For analytic convenience, these populations are often assumed to be unstructured, randomly mating groups of individuals, even when such conditions are rarely met in reality. When assuming discrete populations, it may be difficult to model an individual’s ancestry as arising from a single population; individuals that are modeled as having ancestry from more than one population are often labeled as “admixed” (Falush et al., 2003; Pritchard et al., 2000). Similarly, when a set of individuals is inferred as deriving ancestry from more than one population, that group is sometimes described as an “admixed population,” although individuals within that group may and often do have different degrees of admixture (Lipson et al., 2013; Patterson et al., 2012; Pickrell and Pritchard, 2012).
The concept of admixture carries with it a number of conceptual and interpretive challenges. On one hand, it may be useful for reflecting when an individual or population has lines of ancestry that trace back to multiple distant geographic origins on a recent timescale: for example, in describing individuals from Central and South America whose ancestry 600 years ago traces to individuals mostly living in western Europe, west Africa, and Central/South America. A difficulty, however, is the often-implicit assumption of timescale. All humans are admixed, in the sense of having genetic ancestors that lived in different geographic regions at different times. But not everyone is recently admixed: for some, ancestry lines will trace back to geographically distant ancestors within a few generations whereas for others, the same process occurs on a longer timescale. A further challenge is that admixture is almost always framed in terms of modeling constructs of “source populations,” which may erroneously imply the existence of homogeneous populations in the past. An example of the problem is when describing individuals as admixed between African and European ancestries, when neither label refers to a genetically homogeneous grouping or one that is well defined or even stable over evolutionary time. Thus, an unexamined discussion of admixture can lend itself to racialized thinking. The persistence of such thinking in genetics owes in part to the fact that admixture models have been useful in genetic disease mapping (Chakraborty and Weiss, 1986; Freedman et al., 2006; Pritchard et al., 2000): the models are approximately correct for gene mapping despite the assumptions being precisely wrong.
While genetic ancestry is often the descriptive framing used by researchers to describe individuals or groups in their samples, “most statements about ancestry are really statements about genetic similarity” (Coop, 2022; Mathieson and Scally, 2020). Individuals that share more paths through
the family tree will have accumulated fewer genetic differences between them, and thus on average will have more similar genotypes. This genetic similarity is observable using genotyping and sequencing technologies, and so direct observations of genetic similarity provide an indirect reflection of the ancestral recombination graph and the family trees in which it is embedded (see Figure 2-1 inset). Genetic similarity is thus informative about genetic ancestry and, to a much lesser extent, genealogical ancestry, but the relationship among the three concepts is complicated (Coop, 2022; Coop Lab, 2017a) (see Figure 2-1). Moreover, in practice, researchers are often interested only in genetic similarity—such as when assembling study participants for a genome-wide association study—rather than in ancestry over different time periods, and use ancestry labels only as shorthand, at the risk of considerable confusion.
Geography
In surveys of human genetic diversity, a broad correlation is typically observed between genome-wide genetic similarity and geographic proximity (Rosenberg, 2021). It is no surprise then that ancestry, whether genetic or genealogical, is often expressed in terms of geography. The “origins” or “roots” ascribed to people are routinely denoted by geographic labels, using for example, continents or continental regions, such as European or North African, or political territorial labels at the resolution of a participant’s country of origin, such as Brazilian or Japanese. Geographic proximity is also a crude surrogate for shared environment in studies of ongoing and/or past exposures to environmental factors, whether biotic (such as pathogens) or abiotic (such as solar incidence). Geographic labels thus frequently attribute shared environmental experiences to groups, even when it is the individual exposures that are relevant to understanding traits. In sum, geography is a widely used population descriptor that stands in for more than one concept of human difference. Indeed, part of its methodological appeal may lie in its seeming to obviate making difficult choices, such as if and how to ascribe ethnic labels to individuals.
Yet geography as a population descriptor presents challenges, many of which are similar to those related to genetic ancestry, such as questions of spatial and temporal resolution. For one, geography is a broad concept that can variously be measured by breaking it down into more precise indicators, such as an individual’s birthplace, their current place of residence, or the full trajectory of the locations they have lived. Alternatively, and especially in genetics, the interest may be in the descent-associated notion of geographical ancestry, that is, where an individual’s genealogical ancestors lived at some time point in the past. Geographical ancestry, like genetic ancestry, thus requires having to specify a timescale. A common ap-
proach is to use data from parental or grandparental birthplaces to indicate geographical ancestry one or two generations into the past (Fujimura and Rajagopalan, 2011).
Regardless of the precise geographic location of interest, one common challenge when using geographic population descriptors is how to describe locations. While latitude and longitude can be used in plots and maps, often in a genetics or genomics study, a researcher may wish to find place names to describe a set of geographical locations concisely. This raises the question of how to bin geographic locations together into labels. Various approaches are taken, from using contemporary or past borders of political entities, to referring to features of physical geography. This decision process also raises the issue of spatial resolution. Should the inhabitants of Palermo, for example, be referred to as Sicilian, Italian, Mediterranean, or European?
Of the many options for describing the geography of a sample using population labels, continental-scale summaries of geographic locations are among the most commonly used. Continental-scale population descriptors are problematic for two main reasons: the heterogeneity that exists within continents along nearly any dimension of interest, and the important fact that continent boundaries have no relevance for genetics studies. As examples, neither the classical divide of Asia and Europe in the Bosporus Strait nor separating Asia and Africa at the Suez Canal are meaningful indicators of genetic ancestry. The use of continents as a unit of analysis is arguably part of a larger intellectual tradition of mistakenly viewing continents as homogenous units (Lewis and Wigen, 1997).
Researchers may also face logistical challenges when trying to apply geographic population descriptors. They often have access to only a single self-reported location or one sampling location (like place of residence), but it may not be the location that is most relevant to address the study question (for example, birthplace). Or sometimes a geographic location is imputed from a group label, which inevitably misrepresents geographic origins. For example, for the purposes of carrying out a geographic analysis, a researcher might use the geographic center of France for all individuals labeled as French. In other cases, a geographic location will be intentionally coarse-grained to preserve privacy and hinder de-anonymizing individual-level data (e.g., the use of postal codes to represent geographic locations versus providing precise latitude and longitude). Whether such practices are problematic will depend on the type of analysis and the sensitivity it has to errors.
Finally, the fact that geography is often understood through the filter of geographical ancestry in many areas of genetics may lead to conceptual slippages. For example, in representations of the geography of human genetic variation, samples are often projected on maps according to the location of inferred ancestors rather than on the sampling location, such
as placing the CEPH sample of Utah individuals (CEU) in the category of Central European ancestry, or the Gujaratis sampled in Houston (GIH) in South Asia in figures showing 1000 Genomes data (1000 Genomes Project Consortium, 2015). This slippage may be unimportant for questions related to understanding the genetic history of particular sets of peoples, but it is problematic in research in which geography is a proxy for environmental exposures. As another example, the People of the British Isles project describes the population genetic history of those people of the British Isles with recent ancestors from the British Isles,2 leaving aside for future work understanding the histories of all individuals on the British Isles with recent ancestors from other parts of the world, such as immigrants from South Asia. Such approaches have implications for equity—by focusing on one geographic ancestry, a subset of participants is excluded from analyses. Another related limitation of geography as a population descriptor is its incompleteness. Individuals living within the same geographic area may be highly heterogeneous socially, ethnically, environmentally, and in terms of genetic ancestry. As with the use of genetic ancestry as a population descriptor, careful consideration of the intended uses of a geographic descriptor is essential in every genomics study.
Ethnicity
Ethnicity is often mentioned alongside race without clarifying the relationship between the two. Although the frequent expression race and ethnicity suggests they are separate concepts, it is not unusual for them to be treated as synonyms (or combined in the adjective ethnoracial) (Malinowska and Żuradzki, 2022). Their shared association with descent facilitates this elision, as does the not uncommon desire to find a substitute term for race.
Despite certain resemblances, the terms ethnicity and race have distinct conceptual histories and connotations. While the race notion was formed prior to the twentieth century, the idea of ethnicity was popularized in the early 1900s to capture differences between European-origin groups in the United States (Hattam, 2004). In this context, ethnicity was intended expressly to mean something other than race: linguistic, religious, and other cultural group markers as opposed to the physical traits that supposedly demarcated races.
Ethnicity then can be defined as a sociopolitical system for classifying human beings according to claims of shared heritage that are largely based on perceived cultural similarities (e.g., of language, dress, foodways, reli-
___________________
2 University of Oxford. n.d. People of the British Isles: Population genetics and facial genetics. https://peopleofthebritishisles.web.ox.ac.uk/home (accessed October 19, 2022).
gion) (Cornell and Hartmann, 2007). Its similarity to race stems from the common claims to delineate descent-associated groups, but where racial boundaries have historically been closely tied to supposed biological traits, ethnic groupings are more tightly linked to perceived cultural practices, norms, values, and beliefs (Cornell and Hartmann, 2007).
The frequent conflation of race and ethnicity, however—in the United States and elsewhere (Wimmer, 2013)—means that ethnic groups may be racialized (e.g., ascribed ostensibly racial characteristics, for example in the case of Hispanics/Latinos in the United States), and racial groups can be ethnicized (e.g., Asians being ascribed broad cultural similarities).
The tendency for race and ethnicity—broadly understood as physical and cultural difference—to be intertwined when thinking about the groups that are seen as descending from shared roots means that they are perhaps more distinct in their abstract definitions than they are in everyday language and practice. It is useful, then, to draw a distinction between the separate logics at their core in order to clarify the concepts of difference that are in play when researchers design and assess genomics research.
The type of group that is recognized as an ethnicity varies widely. In the United States, descendants of voluntary immigrants are perhaps the most likely to be labeled as ethnic groups: hence Italian Americans, Korean Americans, and Cuban Americans, for example, are considered ethnic groups (Cornell and Hartmann, 2007). Although African Americans could also be said to constitute a culturally distinct ethnic group, their long and profound history of racialization3 as black has tended to preclude recognition as an ethnic community (Clergé, 2019; Waters, 1990). However, as the African-descent population of the United States becomes increasingly diverse in terms of its migration background (Clergé, 2019; Hamilton, 2019), African Americans are likely to come to be better understood as an ethnic group distinct from, say, Haitian Americans or Nigerian Americans.
Another form of ethnicity that tends not to be recognized as such in the United States is indigeneity; instead, the language of nations and tribes endures to describe the wide variety of native North American groups that are heirs to distinct languages and traditions. The next section explores indigeneity in greater detail, but here the committee wishes to contrast the U.S. tendency to distinguish ethnicity from indigeneity, when outside the United States, ethnicity may often denote groups that are considered indigenous to a given territory. Worldwide, then, ethnicity is not limited to a product of migration.
The United States is also unusual for its official system of ethnoracial classification (OMB Statistical Directive 15), which recognizes only one
___________________
3 Racialization is the process through which groups defined by attributes or characteristics such as skin color, customs, etc. come to be understood as distinct biological entities and human lineages (Hochman, 2019).
ethnicity today: Hispanic or Latino identification. Normally, official questions about ethnicity—notably, as they appear on national censuses—offer respondents a wide range of options from which to choose. The 2021 Canadian census, for example, instructs respondents to indicate as many “ethnic or cultural origins” as applicable and provides a link to a list of more than 500 examples.4 This report does not adopt the OMB’s equation of ethnicity with Hispanicity, but rather it uses the term to denote inclusive and comprehensive classification schema that recognize multiple ethnic groups.
One of the most common ways in which geneticists have incorporated ethnicity in their research is through the inclusion of linguistic information (Barbujani and Sokal, 1990; Cavalli-Sforza et al., 1988; Wang et al., 2007). As Cavalli-Sforza (1997) contended in his study of human genetic history:
Most patterns found in the analysis of human living populations are likely to be consequences of demographic expansions, determined by technological developments affecting food availability, transportation, or military power. During such expansions, both genes and languages are spread to potentially vast areas. In principle, this tends to create a correlation between the respective evolutionary trees. The correlation is usually positive and often remarkably high. (p. 7719)
Linguistics thus often marks the migration of populations across geographic space, as language is something they “carry” with them. As is demonstrated by Western-Hemisphere societies in which such European languages as English and Spanish predominate even though large swathes of their populations trace their roots outside Europe, mother tongues do not necessarily mirror ancestral origins. The reach of language as a reflection of descent is therefore time sensitive, a factor that must be taken into account by researchers who wish to use linguistic data as a measure of ethnicity.
Linguistic patterns are also likely to deviate from other descent-associated population descriptors. But to the extent that a researcher may wish to capture an ethnic sense of belonging; gauge familiarity with the worldviews, norms, and values expressed through a particular language; or link exposure to particular ethnic practices or institutions, language may be of real use as a descent-associated population descriptor. In data collection from living individuals, information on language usage and fluency may be self-reported (or assessed by an external evaluation), whereas the genetic investigations of human evolutionary history that have effectively
___________________
4 See Canada’s long-form 2021 questionnaire at https://www.statcan.gc.ca/en/statistical-programs/instrument/3901_Q2_V6, and its Examples of Ethnic or Cultural Origins at https://www12.statcan.gc.ca/census-recensement/2021/ref/questionnaire/ancestry.cfm (both accessed December 13, 2022).
incorporated language data have tended to rely on classifications developed by linguistic experts (Tishkoff et al., 2009).
In summary, using ethnicity as a population descriptor has the advantage of offering myriad ways to capture it—and thus the challenge of selecting and justifying the choice from among many options. As is true of other descriptors considered in this report, it requires the researcher to reflect carefully and precisely on the mechanism they wish to explore and to collect the data that best measure the variable(s) they have in mind.
Indigeneity
The term Indigenous is generally applied to peoples who retain a historical link to precolonial societies in a given place or region (Martínez Cobo, 1987). Its definition is complex, however, and its use and meaning may differ between local contexts. Like ethnicity, indigeneity carries connotations of descent-associated cultural traditions, and in some places (including the United States), it has been translated into racial categories that emphasize physical traits and political status (Gartner et al., 2021). However, the concept of indigeneity is distinguished from these other population descriptors by its emphasis on the continuity of geographic location over time as well as shared culture, traditions, and other connections (Bello-Bravo, 2019).
The social salience of indigeneity varies a great deal across the globe. The recognition of indigeneity seems to be more common in regions and countries outside Europe that have been subjected to Europeans’ relatively recent colonial expansions. Accordingly, the label Indigenous is rarely applied to populations within Europe itself—that is, the Sámi are the only Indigenous people recognized within the European Union (The Swedish Development Forum, 2018). Consistent with this observation, a study of censuses conducted around the world between 1995 and 2004 found that no European nation fielded a question on Indigenous or tribal status, compared to 67 percent of those in South America, where it was most frequent (Morning, 2008).
There are also striking differences in the terminology used to refer to Indigenous peoples around the world (Bartlett et al., 2007). Variants of the term “Indian” (e.g., American Indian, indio) continue to be widely used across the Americas to refer to Indigenous ancestries from the Western Hemisphere, in the biomedical literature as well as in popular parlance (Benedet et al., 2012; Egan et al., 2017; Goicoechea et al., 2001; Guardado-Estrada et al., 2009; Marca-Ysabel et al., 2021; Spangenberg et al., 2021). Indigenous peoples in Australia are often referred to as “aboriginal” (Malaspinas et al., 2016; Nasir et al., 2022; Rasmussen et al., 2011), whereas New Zealand is more specific in recognizing the Māori as Indigenous and distinguishing them from other Polynesian populations in the West Pacific,
which are often referred to with geographic labels such as Tongan, Samoan, or Cook Islanders (Tätte et al., 2022; Umaefulam et al., 2022; Wang et al., 2022). In short, the nomenclature associated with indigeneity goes from very broad and homogenizing groupings—such as native or Indian—that primarily serve to distinguish them from the non-Indigenous, to much narrower, more geographically delimited ethnic identifiers such as Māori or Lenape (Peters, 2011). Depending on the granularity of these descriptors, genomics studies can have very different resolutions and interpretations, and, therefore, comparability to other studies.
In addition to its association with geographical rootedness over time, indigeneity often carries a connotation of “purity” or absence of admixture with non-Indigenous peoples. In such interpretations, indigeneity functions as a racial category, complete with the kind of references to blood quantum (as in half-blood or full-blood) that played an important role in the development of the race concept (Gartner et al., 2021). Today, American Indian/Alaska Native peoples in the United States often have to prove and substantiate their tribal enrollment status through blood quantum in order to qualify for health and other governmental benefits specified by treaty obligations (Spruhan, 2017).
Many countries have adopted (or inherited) classification systems that aim to discern different degrees of mixing between European and Indigenous populations during and since colonial times; the castas taxonomies of Spanish colonies in the Americas offer a prime example (Katzew, 2004; Moreno Navarro, 1973). This is especially evident in areas where admixture was believed to lead to the “whitening”—and ultimate disappearance—of Indigenous peoples, releasing their lands for European occupation (Wolfe, 2001).
Such is the case of the term mestizo (Spanish for admixed), which is used across Spanish-speaking Latin America, including in the biomedical and genetics literature (Silva-Zolezzi et al., 2009; Wang et al., 2008). In colonial times, mestizo was used to refer to the offspring of Spaniards with Indigenous partners, and over time it has been generalized to distinguish almost everyone who is not Indigenous (Rodriguez Mega, 2021). Thus, as with other group labels, mestizo is inaccurate because it socially homogenizes a genetically heterogeneous group of people.
The U.S. terms American Indian and Alaska Native are broadly used to describe the more than 574 federally recognized tribal nations (excluding the state-recognized or unrecognized tribes), each of which exists as a sovereign entity with its own distinct culture and language (NCAI, 2020). Tribal nations have final determination of who they designate as tribal members, sometimes determined through direct familial ties, descent, or through a minimum blood quantum value (U.S. Department of the Interior, n.d.). The terms Hispanic and Latino also amount to broad single descriptors applied to a heterogeneous and diverse group of populations with varying degrees
of recent African, European, and Indigenous American genetic ancestries (Bryc et al., 2010, 2015). In Canada, the term Métis refers to individuals of mixed recent European (primarily French) and Indigenous genetic ancestry, distinguishing them from First Nations and Inuit Indigenous peoples (Bartlett et al., 2007).
In other regions of the world, such as North Africa, the Middle East, and Central Asia, there is not a generalized term that aims to distinguish Indigenous from admixed peoples. Despite the high degree of admixture among North Africans, primarily involving Indigenous North African ancestry, near Eastern Arab ancestry, and European ancestry, or across Asia between Turkic and Mongolian groups and earlier inhabitants, Indigenous ancestries tend to be described in the literature by either specific geographic terms (e.g., Maghrebi, Qatari) or specific ethnicities (e.g., Berber, Saharawi, Bedouin, Hazara, Azeri) (Botigué et al., 2013; Henn et al., 2012; Koshy et al., 2017; Rodriguez-Flores et al., 2016). In these regions where the degree of European ancestry is not a focal point for describing admixture, indigeneity is not perceived as distinct from or inconsistent with admixture. In self-identified Berber as well as Arab communities in North Africa, local populations are often described as mixtures of various Indigenous components (Arauna et al., 2017; Lucas-Sánchez et al., 2021).
Despite the variety of conceptual and terminological approaches to indigeneity found around the world, a common theme is that of the sensitivity of nomenclature and classification. This is not unique to indigeneity; other population descriptors raise similar issues. Yet labels of Indigenous clearly pose myriad difficulties. For one thing, the widely used term Indian for the original inhabitants of the Western Hemisphere is a well-known misnomer, even as it has been broadly used by both Indigenous and non-Indigenous Americans (Snipp and National Committee for Research on the 1980 Census, 1989; Snipp, 2002). In Australia, the term “aboriginal” has come under scrutiny for its association with a long and ongoing history of discrimination as well as controversial research practices (Kowal et al., 2012). And the categories meant to distinguish admixed populations from supposedly “pure” Indigenous ones are, as shown previously, overly broad and homogenizing.
These challenges and others make clear that researchers must employ population labels with great care, paying attention to their connotations (historical and contemporary), understanding the terms preferred by the groups in question, and whenever possible, engaging with the communities affected to arrive at descriptors that respect group self-understandings and self-identification. An example is the effort led by Māori researchers in biobanking and genetics research in New Zealand; they worked to incorporate Māori perspectives from the outset to provide a framework for global consideration of Indigenous knowledge in describing and engaging with local communities (Beaton et al., 2017; Kathlene et al., 2022).
Race
The concept of race has roots in the European project of settler-colonialism and slavery and emerged gradually over the sixteenth to nineteenth centuries (Keel, 2018; Mahmud, 1999). Race as it is understood today derives from historical and modern forms of racism and while racial thinking shares certain elements in common with other classification systems based on, say, religion or color, race differs in the specific configuration of assumptions made about the nature of human difference. Smedley and Smedley (2012) identify five ideological ingredients of the racial worldview as it developed in the United States:
- All humans can be categorized into a universal set of self-evident, discrete, exclusive biological groups.
- These groups are ranked hierarchically.
- Surface-level traits used to demarcate races, such as skin color, reflect deeper, essential differences in cognitive, cultural, or moral attributes.
- These attributes are heritable, such that essential differences remain stable over time.
- These distinct groups exist in nature or are the product of divine creation.
These assumptions are not unique to the United States; they also characterize forms of racial classification that crystallized in other contexts of European settler-colonialism by the nineteenth century, such as in South America or Australia (Wolfe, 2016). Across these varied settings, race is fundamentally a sociopolitically constructed system for classifying and ranking human beings according to subjective beliefs about shared ancestry and innate biological similarities.
Race was integral to Enlightenment-era understandings of human nature and helped to resolve a contradiction reflected in two competing elements of political and social discourse (Fredrickson, 2002; Keel, 2018; Roberts, 2011). On the one hand, Western political theorists and actors championed the principles of liberty, equality, rights, and happiness. On the other hand, however, their societies were deeply enmeshed in colonialism and enslavement, which were enormously profitable for elites but the antithesis of democratic values. Theories of race resolved the contradiction between the discourse of liberty and the reality of slavery by inventing hierarchical taxonomies of fixed subdivisions, or types, of humankind, each with different capacities and natural stations in life. The supposedly superior and inferior races that Linnaeus and other early scientists described were not thought to possess the same rights, privileges, or even humanity—
thus seemingly justifying the freedom of some and the subjugation of others (Guyatt, 2016; Morgan, 1975; Wolfe, 2016).
The history of race, science, and society is important for contemporary genetics and genomics researchers to understand, because it illustrates that race emerged as a political project well before the development of population genetics or the evolutionary theory that sustains it. However, including that history here in sufficient detail is beyond the scope of this report and outside the committee’s statement of task. For a deeper understanding of race and racism and their impacts on science and society, begin with the references in Box 1-1.
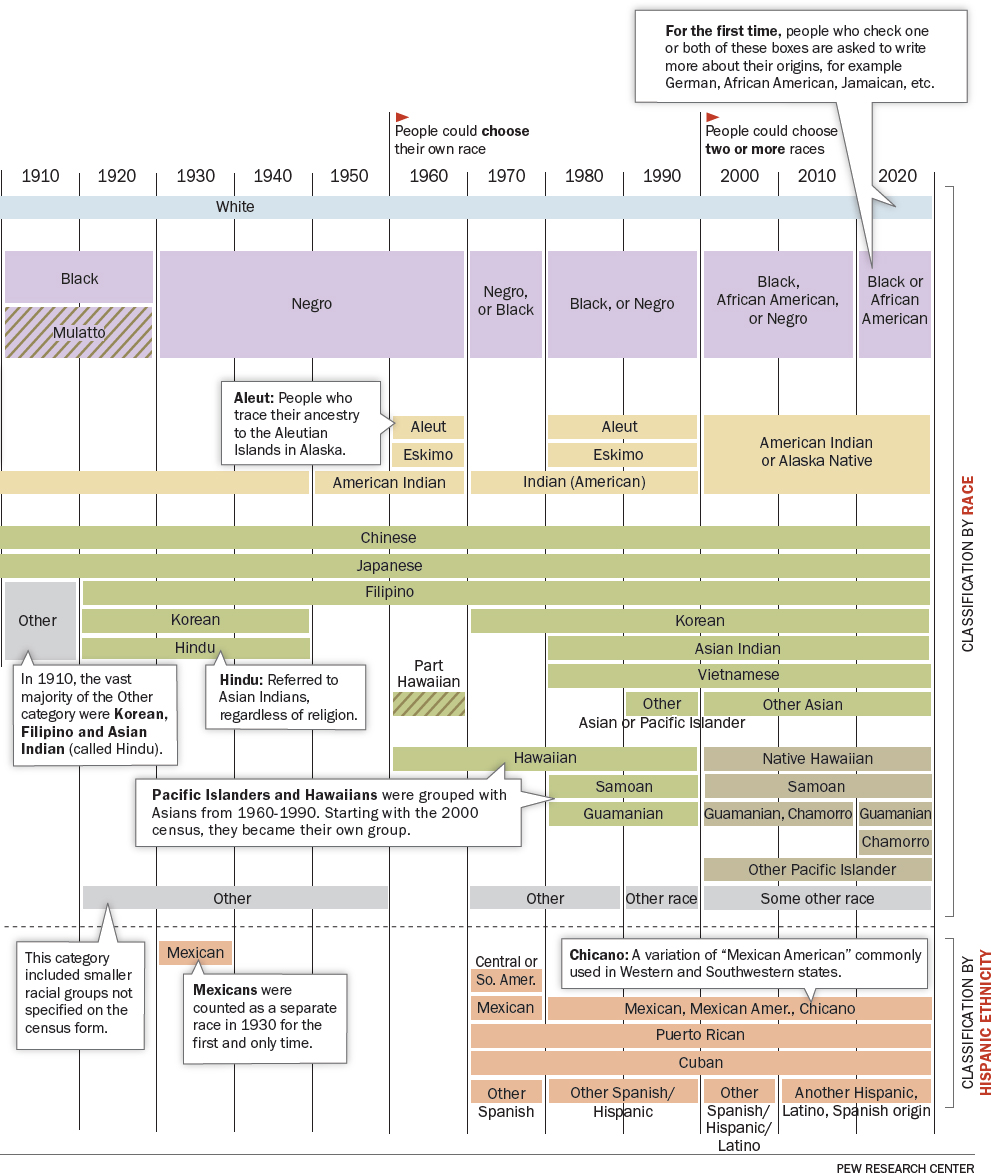
The social nature of race is especially evident in the United States, where it was not only embedded in early governing structures like the Constitution or naturalization law, but where racial categories have constantly evolved. The list of racial groups on the U.S. Census, for example, has changed nearly every decade since the first enumeration in 1790 (Figure 2-2), with categories like “mulatto,” “Mexican,” and “Hindu” appearing and disappearing (Lee, 1993; Prewitt, 2005). In the early twentieth century, Italians, Jews, and the Irish were popularly considered to be racial groups, distinct from the white majority with Northern European ancestry (Jacobson, 1998). There have been profound disagreements in both the demarcations between races and the meaning of race itself. These disagreements have never been only a battle of abstract ideas—or of scientific precision. Racial thinking emerges from historical and contemporary forms of racism and retains its force because of the way it buttresses political, economic, and social structures.
In addition to the dynamic nature of how race is conceptualized, race is also a complex population descriptor because it can be interpreted and measured in many ways. As sociologist Wendy Roth (2016) puts it, race is “multidimensional,” and she identifies six broad dimensions of race:
- “Racial identity” refers to an individual’s self-identification;
- “Racial self-classification” describes how an individual reports their race in formal settings with a limited menu of options, like on a census form;
- “Observed race” is the race that observers ascribe to an individual, based on appearance or behavior;
- “Reflected race” is what an individual believes others take them to be;
- “Phenotype” corresponds to visible physical traits that are thought to be indicators of racial membership; and
- “Racial ancestry” captures the racial identities ascribed to one’s ancestors (as reported by family lore, for example, or inferred by genetic genealogy companies).
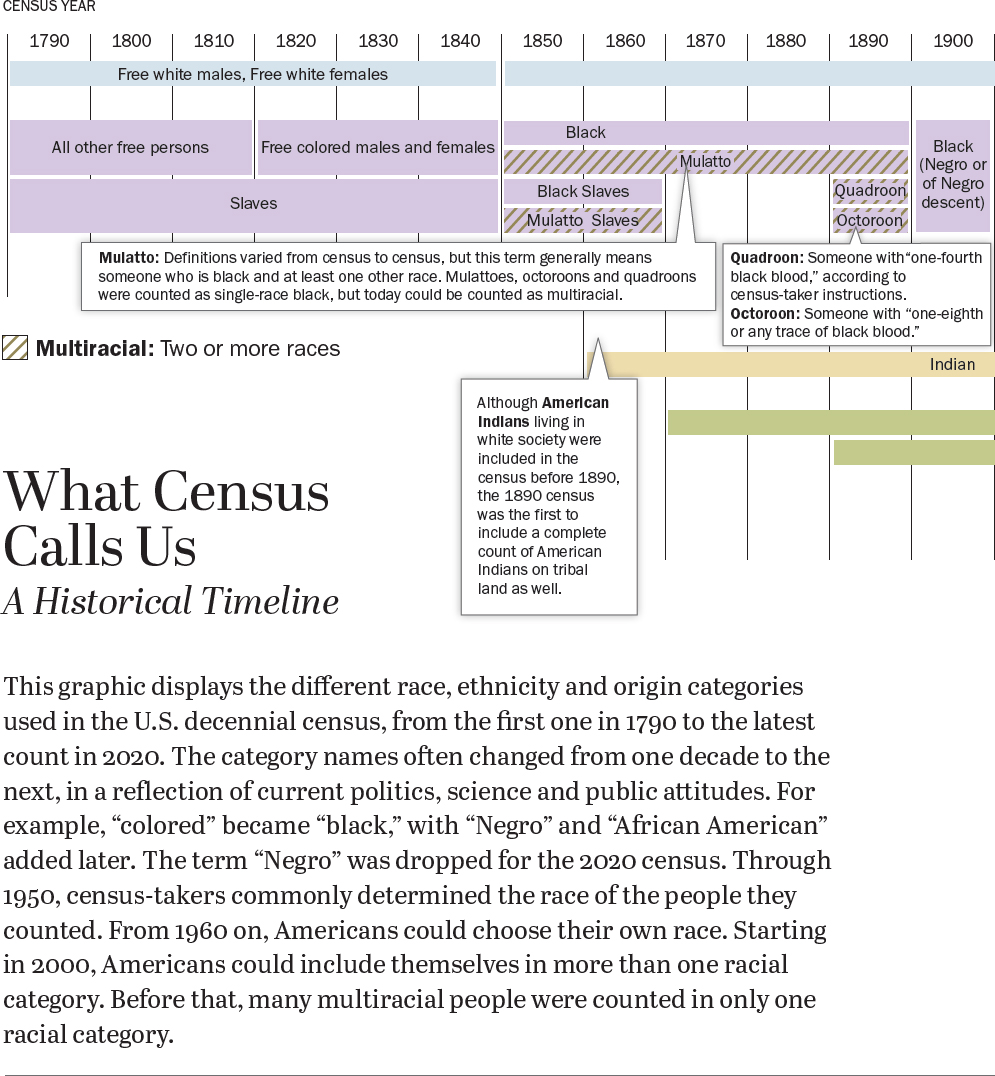
SOURCE: Pew Research Center, https://www.pewresearch.org/wp-content/uploads/2020/02/PH_15.06.11_MultiRacial-Timeline.pdf. Courtesy of Pew Research Center.
As research on individuals’ “contested identities” shows, these dimensions are not necessarily aligned with each other (Vargas and Kingsbury, 2016). An individual who self-identifies as Asian and white might check off “white” alone on a survey yet be perceived as Asian by an observer. In the rare cases where race is used as a proxy for the experience of racism in health disparities studies and its genetic contributions, researchers should carefully consider which dimension of racial identity is best suited to their study objective, be mindful of which dimension of race their data actually measures, and be transparent in specifying it.
As these dimensions of race suggest, it is also important to indicate the source of racial data: are the data based on individuals’ self-reports of their racial identity? Or perhaps an external observers’ impressions of their phenotype? Equally important is acknowledging the fluidity of racial identity, making it crucial to indicate the point in time at which the data were collected. Liebler et al. (2017) determined that nearly 10 million individuals changed their racial self-classification on the U.S. census between 2000 and 2010, making clear that race should not be presumed to be a stable personal characteristic over time. Andrew Penner and Aliya Saperstein show, moreover, that not only are such fluctuating racial categorizations true of observers’ reports as well, but that these changes in racial identification—whether individually chosen or externally assessed—are associated with the social status of the individual whose race is being identified (Penner and Saperstein, 2018; Saperstein and Penner, 2012).
For all these reasons (and more—see Box 1-1 and Chapter 1), race makes for a poor proxy of human biological variation. However, it may be a useful population descriptor for researchers who wish to measure a consequential form of social status and affiliation, in the absence of other data (notably, the case where race may be a proxy for the experience of racism in health disparities studies, see Chapter 5). As W.E.B. Du Bois (1923) famously put it, “the black man is a person who must ride ‘Jim Crow’ in Georgia.” This definition tells us that while race lacks the precision, rigor, stability, and objectivity one might hope for in a population descriptor, it is a social construct that speaks volumes about its time and place. Applied to any specific human being, however, it is a blunt instrument with which to infer individual social standing, economic status, or experiences of discrimination (Monk, 2022). For this reason, the committee urges researchers to think deeply about the specific experiences, characteristics, or outcomes they wish to investigate, and then consider searching for or constructing measures that capture these more directly and precisely than race ever can.
THE IMPORTANCE OF ENVIRONMENTAL FACTORS IN GENETICS AND GENOMICS RESEARCH
This final section of the chapter touches on three topics. First, the genome does not operate in a physiological vacuum. An organism’s internal milieu and the external world it inhabits interact with the genome to affect phenotypic expression and variability (Alon, 2006). Second, to better understand the interplay of genes and the environment in humans, descent-associated population descriptors should frequently be accompanied by measures of the population’s relevant environment(s) in genetics studies. Finally, researchers are increasing the understanding of environment effects on phenotypes through studies of the epigenome—the sum total of all chemical modifications that occur on the DNA sequence of the genome to modulate the activity of genes (Felsenfeld, 2015). The epigenome can thus be viewed as one liaison between the environment and the genome. Hence, it will be increasingly valuable to incorporate epigenetic measurements into studies of human genetic variation.
Genes, Environments, and their Interactions
It has long been recognized that phenotypes are the result of interplay between an individual’s genome and their environment (Li et al., 2018; Seabrook and Avison, 2010; Strickberger, 1985). Breeders and farmers have used this knowledge for centuries to select for and breed crops and domestic animals with desirable traits. Genetics studies have shown us that this interplay is complex, dynamic, and often hard to untangle. A gene’s expression depends on a range of factors from an organism’s internal physiologic state to signals received about the immediate external milieu, as well as on the stage of the cell cycle and the state of the epigenome (Boyce et al., 2020). Importantly, the genome interacts with the environment throughout an individual’s lifetime to form an aggregate phenotype.
Some of the earliest genetics studies in the fruit fly Drosophila, corn, and the bread mold Neurospora examined the effects of the external environment such as temperature, light, and nutrition, respectively, on the occurrence of mutations (genetic variants) (Strickberger, 1985). Other studies, including in humans, examined the effects of the internal environment as measured by age and sex (Strickberger, 1985). This body of literature suggested two new insights: (1) some genotypes exhibit differential expression depending on the environment (a gene–environment interaction) (Gibson, 2008; Smith and Kruglyak, 2008), and (2) relatives share not only genes but also an environment, called the shared environment, which leads to a correlation of genetic backgrounds with environments (Mayhew and Meyre, 2017). Thus, phenotypic similarity of two individuals depends on their genetic variation profiles (genotypes), their shared and unique environments,
as well as the resulting interactions between each individual’s genome and their environment (see the effects on stature in Jelenkovic et al., 2016).
So, what might be the consequence of ignoring the environment? When the environmental effects vary, then failing to take them into account is at best incomplete and more likely misleading. This might arise in two ways. First, the environment can vary independent of genes, as most genetics studies assume. Not studying the environment in this case means that the analysis is incomplete, and further, generalizing the effects of the genetics that were uncovered to new environmental settings is problematic and can be error prone. Second, the environment covaries with genetics so that interpreting the results of phenotypic similarity in terms of genetics alone would simply be misleading and can overemphasize its effects (Mayhew and Meyre, 2017).
Defining the Environment
Settling on a definition of the environment is tricky, because it is contextual, varies among scientific disciplines, and necessarily groups highly diverse concepts under one umbrella term. As Robette (2022) observes,
One might wonder what effects the use of the term environment without epistemological scrutiny has on the construction of sociological objects. In the additive polygenic model, the environment is anything that is not genetic. In the context of epidemiology, the environment refers mainly to risk factors (diet, pollution, etc.). For sociogenomicists, the environment seems synonymous with the social. The use of the term without conceptual work thus leads to the grouping of highly diverse processes under the same term. (pp. 198-199)
For any given study, a researcher has to identify which environmental factors to examine and how to measure them accurately, as with any descent-associated population descriptors.
For any given trait, only a limited number of environmental factors are likely at play, some of which can be gleaned from the epidemiologic literature. In the absence of directly measuring environmental factors, epidemiologic and genetics studies sometimes use race or ethnicity as a proxy for measures such as cultural beliefs or shared environment (Benmarhnia et al., 2021; Duello et al., 2021; Martinez et al., 2022).
Social context, an attribute of the environment, influences behavior and interacts dynamically with biology, including genetics, throughout the life course to affect human health (Glass and McAtee, 2006). The social context or social environment can be defined as the society and physical surroundings and conditions in which a person lives, works, and interacts with others. It can be measured in a variety of ways, capturing multiple dimensions
(Boardman et al., 2012; Duncan and Kawachi, 2018; Williams et al., 2019). It can be objective or perceived/self-reported and can vary in spatial scale depending on the boundaries set by geography and/or the researcher (Diez Roux and Mair, 2010; Duncan and Kawachi, 2018). Heuristically, the social environment can be classified into two broad categories, which are not necessarily mutually exclusive and may interact with each other:
- Social conditions: examples include social support, social networks, income inequality, deprivation and poverty, education, social capital, segregation, discrimination, demographic composition, and violence.
- Built or physical environment: examples include land use, green spaces, pollution, traffic noise, and population density.
Structural racism is a system of oppression that shapes the conditions and resources of the environment, such as those listed in the two categories above. Researchers are starting to measure structural racism using a combination of variables at the state and Metropolitan Statistical Area levels (Hardeman et al., 2022; Homan et al., 2021; Krieger, 2021). The concentration of racialized and ethnic groups in the United States is unevenly distributed owing to sociopolitical processes such as segregation and poverty (Williams et al., 2019); therefore, to avoid biologizing race and ethnicity, it is especially important to directly quantify the environment when conducting research aimed at understanding phenotypes that result from both genetic and environmental effects. This is necessary because genetic variation and environmental effects can be correlated or interact. Correlation occurs when genotypes or presumed ancestries are not randomly distributed across environments (e.g., ancestries of children exposed to lead-based paint in their homes), and interaction happens when genetic effects depend on the environment (e.g., phenylketonuria occurs only with phenylalanine in the diet). Thus, ancestry-specific or ancestry-enriched markers may show an excess with the developmental growth outcomes measured from lead exposure, but these are hardly the biological etiologic factors geneticists wish to discover. Some genetics studies can correct this effect in part using genetic markers, but the etiologic factor is lead. Yet in both examples, recognition of the environmental agent is central to understanding the outcomes and phenotypes that result from both genetic and environmental effects. Furthermore, for example, residential segregation and community demographic composition are considered in multilevel analysis to capture social exposures that may be embodied and affect an individual’s biology (e.g., psychosocial stress associated with living in a poor neighborhood may lead to hypertension) (Arcaya and Schnake-Mahl, 2017; Kramer and Hogue, 2009; Parks, 2016; Williams and Collins, 2001).
Much of the evidence on the association between environmental factors and health draws on studies centered on spatially defined geographic areas. Some of the spatially defined areas are viewed as proxies for neighborhood. However, neighborhood boundaries are often inferred from administrative areas (e.g., census-defined geographic areas such as tracts, block groups, and zip codes) rather than neighborhoods defined more in terms of social boundaries or even ecological boundaries. Other spatially defined boundaries are also possible based on proximity to the individual (e.g., the density of violence), and persons often cross boundaries in their daily lives, bringing into play multiple types of environmental influences.
The preceding discussion emphasizes that although researchers can qualitatively and quantitatively measure some aspects of the environment related to individual research participants, this cannot, with today’s knowledge, be complete. Unlike the genome, which is large yet finite and can be studied completely (at some level of resolution), an exhaustive characterization of environmental effects is not feasible. It is difficult to determine the critical period, the dose-response, or the specific environmental factor affecting the human genome. In addition, most environmental factors are correlated with each other as they may be determined by the same upstream cause, e.g., structural racism. These limitations represent key challenges for the interpretation of genetics and genomics studies but compel us to begin to make meaningful strides toward defining the roles of selected and plausible common and unique environments in the traits studied, though admittedly, this is a much harder task for some phenotypes than for others. Consequently, there may be merit to studies of the epigenome that can capture some of the cellular consequences of environmental effects even when the specific environment is unknown.
Epigenetics as Environmental Sensors
Environmental exposures can change gene expression and a consequent phenotype. For example, a remarkable study seeking the molecular causes of thalidomide embryopathy demonstrated that thalidomide phenocopies Mendelian genetic syndromes (Donovan et al., 2018). Starting with a previous observation that genetic mutations in the transcription factor SALL4 led to Duane Radial Ray syndrome and Holt-Oram syndrome, the authors showed that thalidomide directly degrades the SALL4 protein. There are other compelling examples, such as induced nicotinamide adenine dinucleotide (NAD) deficiency in the offspring of mice. Restricting the NAD precursors tryptophan and vitamin B3 in the maternal diet during pregnancy led to an increased frequency of miscarriages and congenital abnormalities in their offspring (Cuny et al., 2020). Interestingly, recessive homozygous variants in HAAO or KYNU, two genes of the NAD synthesis pathway, cause
similar phenotypes in humans and mice, whereas a single copy of these variants that does not lead to an abnormal phenotype alone can be made to do so when combined with maternal diet-mediated NAD deficiency. The molecular basis and the developmental pathways involved are well defined, as they are for the etiology of congenital scoliosis (Sparrow et al., 2012). Taken together, these and other studies imply that (1) phenotype similarity between two individuals can arise from genes only, environments only, genes in one and the environment in the other, or from the interactions between genes and the environment in the same individual, and (2) despite the different proximal causes, the distal molecular causes of the effects of environmental perturbations and those of genetic variants can converge on the same molecular process and the same molecules, at some rate-limiting step (Chakravarti and Turner, 2016).
The question is how genetic and environmental effects—two different proximal causes—can mediate the same outcome. The answer often involves the epigenome. There are two major paths to these changes. In the first, the genome sequence is chemically modified by DNA methylation, which alters the expression of genes (Allis et al., 2015). This process is both genetically programmed and affected by external environment factors, such as diet (Dolinoy, 2008). In the second, the expression of each gene is exquisitely controlled by RNA and protein molecules such that its transcription into RNA and translation into protein is monitored and that information is fed back to regulate the gene’s level and timing of expression (Davidson, 2010). This regulatory machinery, also epigenetic, involves chromatin, transcription factors, and enhancer sequences that are bound by the transcription factors (Allis et al., 2015). Both types of epigenetic machinery are important determinants of phenotype expression (Chakravarti and Turner, 2016). This molecular machinery can today be defined for each human cell type and tissue, and its variation studied in differential traits (e.g., normotensives versus hypertensives). Consequently, a person’s epigenome appears to be functioning as a possible sensor of the environment in which they reside.
An important unanswered question that arises from the above discussion is the time period over which the effects of an exposure can be detected in the epigenome. Some epigenetic modifications, such as parent-of-origin-specific genomic imprinting, are genetically programmed and have lifelong effects on gene and organismal physiology. Others, such as the effects of dietary manipulations, particularly those that affect DNA methylation, are short term, though when they occur early in development can have long-term phenotypic effects (Weyrich et al., 2018). In general, however, the timing, maintenance, or erasure of epigenetic effects on the human genome and phenotype is a largely unexplored area of study.
Epigenetics holds some promise for understanding the consequences of the role of some environments over the life course, especially windows of vulnerability such as childhood that may have long-term consequences for traits decades later in life (Shanahan and Hofer, 2011). This has given rise to the evolving area of social epigenetics (Evans et al., 2021; Martin et al., 2022). However, there needs to be further research on the causal relationship between the external and internal pathways and how these lead to phenotypic change.
Quantifying Environments at the Individual Level
Environmental variables used in research studies can be qualitative or quantitative. There is merit to variables that are quantitative or metrical and show a monotonic relationship with a phenotype of interest (e.g., blood pressure) or disease risk (e.g., hypertension-induced target organ damage). Whenever feasible, these should be measured at the individual research participant level similar to how genotypes are measured. However, this is a highly challenging endeavor. The exposures relevant to different phenotypes may be during development or in years past and, thus, almost impossible to measure, although there may be an epigenetic imprint of such exposures. Even when an exposure is proximal, such as in the survivors of the atomic bombings in Hiroshima and Nagasaki, individual-level radiation exposure was very challenging to quantify because it depended on distance from the blast site, whether the individual was inside or outside, and the type of dwelling in which they resided, among other factors (NRC, 1991). Thus, in many investigations, the only plausible contrast may be between cases and controls, rather than exposed versus unexposed, with no resolution at the level of the individual.
The interest on the part of such funding agencies as the National Institute of Environmental Health Sciences to further develop “exposome” science, preferably inclusive of the “socioexposome,” is laudatory.5 The interest in obtaining multiscale measures of the geospatial environment for large-scale population studies is of great benefit to genetics (Cui et al., 2022). Like the Human Genome Project, which had a staged approach to developing higher resolution and more accurate maps of the human genome, similar efforts in developing individual-level and dynamic exposome measures are necessary to understand the full spectrum of genes, environments, and their interactions, both in individuals and in populations.
___________________
5 https://www.niehs.nih.gov/research/supported/exposure/bio/index.cfm (accessed January 3, 2023).
REFERENCES
1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526:68-74.
Allis, C. D., M. L. Caparros, T. Jenuwein, and D. Reinberg. 2015. Epigenetics. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Alon, U. 2006. An introduction to systems biology: Design principles of biological circuits. 1st ed. London: CRC Press.
Arauna, L. R., J. Mendoza-Revilla, A. Mas-Sandoval, H. Izaabel, A. Bekada, S. Benhamamouch, K. Fadhlaoui-Zid, P. Zalloua, G. Hellenthal, and D. Comas. 2017. Recent historical migrations have shaped the gene pool of Arabs and Berbers in North Africa. Molecular Biology and Evolution 34(2):318-329.
Arcaya, M. C., and A. Schnake-Mahl. 2017. Health in the segregated city. In The dream revisited. New York: NYU Furman Center.
Barbujani, G., and R. R. Sokal. 1990. Zones of sharp genetic change in Europe are also linguistic boundaries. Proceedings of the National Academy of Sciences 87(5):1816-1819.
Bartlett, J. G., L. Madariaga-Vignudo, J. D. O’Neil, and H. V. Kuhnlein. 2007. Identifying indigenous peoples for health research in a global context: A review of perspectives and challenges. International Journal of Circumpolar Health 66(4):287-370.
Beaton, A., M. Hudson, M. Milne, R. V. Port, K. Russell, B. Smith, V. Toki, L. Uerata, P. Wilcox, K. Bartholomew, and H. Wihongi. 2017. Engaging Māori in biobanking and genomic research: A model for biobanks to guide culturally informed governance, operational, and community engagement activities. Genetics and Medicine 19(3):345-351.
Bello-Bravo, J. 2019. When is indigeneity: Closing a legal and sociocultural gap in a contested domestic/international term. AlterNative: An International Journal of Indigenous Peoples 15(2):111-120.
Benedet, A. L., C. F. Moraes, E. F. Camargos, L. F. Oliveira, V. C. Souza, T. C. Lins, A. D. Henriques, D. G. Carmo, W. Machado-Silva, C. N. Araújo, C. Córdova, R. W. Pereira, and O. T. Nóbrega. 2012. Amerindian genetic ancestry protects against Alzheimer’s disease. Dementia and Geriatric Cognitive Disorders 33(5):311-317.
Benmarhnia, T., A. Hajat, and J. S. Kaufman. 2021. Inferential challenges when assessing racial/ethnic health disparities in environmental research. Environmental Health 20(1):7.
Boardman, J. D., E. Roettger, B. W. Domingue, M. B. McQueen, B. C. Haberstick, and K. M. Harris. 2012. Gene-environment interactions related to body mass: School policies and social context as environmental moderators. Journal of Theoretical Politics 24(3):370-388.
Botigué, L. R., B. M. Henn, S. Gravel, B. K. Maples, C. R. Gignoux, E. Corona, G. Atzmon, E. Burns, H. Ostrer, C. Flores, J. Bertranpetit, D. Comas, and C. D. Bustamante. 2013. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proceedings of the National Academy of Sciences 110(29):11791-11796.
Boyce, W. T., M. B. Sokolowski, and G. E. Robinson. 2020. Genes and environments, development and time. Proceedings of the National Academy of Sciences 117(38):23235-23241.
Bryc, K., C. Velez, T. Karafet, A. Moreno-Estrada, A. Reynolds, A. Auton, M. Hammer, C. D. Bustamante, and H. Ostrer. 2010. Colloquium paper: Genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proceedings of the National Academy of Sciences 107(Suppl 2):8954-8961.
Bryc, K., E. Y. Durand, J. M. Macpherson, D. Reich, and J. L. Mountain. 2015. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. American Journal of Human Genetics 96(1):37-53.
Cavalli-Sforza, L. L. 1997. Genes, peoples, and languages. Proceedings of the National Academy of Sciences 94(15):7719-7724.
Cavalli-Sforza, L. L., A. Piazza, P. Menozzi, and J. Mountain. 1988. Reconstruction of human evolution: Bringing together genetic, archaeological, and linguistic data. Proceedings of the National Academy of Sciences 85(16):6002-6006.
Chakraborty, R., and K. M. Weiss. 1986. Frequencies of complex diseases in hybrid populations. American Journal of Physical Anthropology 70(4):489-503.
Chakravarti, A., and T. N. Turner. 2016. Revealing rate-limiting steps in complex disease biology: The crucial importance of studying rare, extreme-phenotype families. Bioessays 38(6):578-586.
Chandra, K. 2012. Constructivist theories of ethnic politics. Oxford University Press.
Clergé, O. 2019. The new noir: Race, identity, and diaspora in black suburbia. Berkeley, CA: University of California Press.
Coop, G. 2023. Genetic similarity versus genetic ancestry groups as sample descriptors in human genetics. arXiv (preprint).
Coop Lab. 2017a. Our vast, shared family tree. https://gcbias.org/2017/11/20/our-vast-shared-family-tree/ (accessed November 7, 2022).
Coop Lab. 2017b. Where did your genetic ancestors come from? https://gcbias.org/2017/12/19/1628/ (accessed October 20, 2022).
Cornell, S., and D. Hartmann. 2007. Ethnicity and race: Making identities in a changing world. Thousand Oaks, CA: Pine Forks Press.
Cui, Y., K. M. Eccles, R. K. Kwok, B. R. Joubert, K. P. Messier, and D. M. Balshaw. 2022. Integrating multiscale geospatial environmental data into large population health studies: Challenges and opportunities. Toxics 10(7):403.
Cuny, H., M. Rapadas, J. Gereis, E. M. M. A. Martin, R. B. Kirk, H. Shi, and S. L. Dunwoodie. 2020. NAD deficiency due to environmental factors or gene-environment interactions causes congenital malformations and miscarriage in mice. Proceedings of the National Academy of Sciences 117(7):3738-3747.
Davidson, E. H. 2010. Emerging properties of animal gene regulatory networks. Nature 468:911-920.
Diez Roux, A. V., and C. Mair. 2010. Neighborhoods and health. Annals of the New York Academy of Sciences 1186:125-145.
Dolinoy, D. C. 2008. The agouti mouse model: An epigenetic biosensor for nutritional and environmental alterations on the fetal epigenome. Nutrition Review 66(Suppl 1):S7-S11.
Donnelly, K. P. 1983. The probability that related individuals share some section of genome identical by descent. Theoretical Population Biology 23(1):34-63.
Donovan, K. A., J. An, R. P. Nowak, J. C. Yuan, E. C. Fink, B. C. Berry, B. L. Ebert, and E. S. Fischer. 2018. Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane-radial ray syndrome. eLife 7:e38430.
Du Bois, W. E. B. 1923. The superior race (an essay). The Smart Set: A Magazine of Cleverness 70(4):55-60.
Duello, T. M., S. Rivedal, C. Wickland, and A. Weller. 2021. Race and genetics versus ‘race’ in genetics: A systematic review of the use of African ancestry in genetic studies. Evolution, Medicine, and Public Health 9(1):232-245.
Duncan, D. T., and I. Kawachi. 2018. Neighborhoods and health. 2nd ed. Oxford, UK: Oxford University Press.
Egan, K. J., H. Campos Santos, F. Beijamini, N. E. Duarte, A. R. V. R. Horimoto, T. P. Taporoski, H. Vallada, A. B. Negrão, J. E. Krieger, M. Pedrazzoli, K. L. Knutson, A. C. Pereira, and M. von Schantz. 2017. Amerindian (but not African or European) ancestry is significantly associated with diurnal preference within an admixed Brazilian population. Chronobiology International 34(2):269-272.
Evans, L., M. Engelman, A. Mikulas, and K. Malecki. 2021. How are social determinants of health integrated into epigenetic research? A systematic review. Social Science in Medicine 273:113738.
Falush, D., M. Stephens, and J. K. Pritchard. 2003. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164(4):1567-1587.
Felsenfeld, G. 2015. A brief history of epigenetics. In Epigenetics. 2nd ed., edited by C. D. Allis, M. L. Caparros, T. Jenuwein, and D. Reinberg. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. Pp. 1-10.
Frederickson, G. M. 2002. Racism: A short history. Princeton, NJ: Princeton University Press.
Freedman, M. L., C. A. Haiman, N. Patterson, G. J. McDonald, A. Tandon, A. Waliszewska, K. Penney, R. G. Steen, K. Ardlie, E. M. John, I. Oakley-Girvan, A. S. Whittemore, K. A. Cooney, S. A. Ingles, D. Altshuler, B. E. Henderson, and D. Reich. 2006. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proceedings of the National Academy of Sciences 103(38):14068-14073.
Fujimura, J. H., and R. Rajagopalan. 2011. Different differences: The use of ‘genetic ancestry’ versus race in biomedical human genetic research. Social Studies of Science 41(1):5-30.
Gartner, D. R., R. E. Wilbur, and M. L. McCoy. 2021. “American Indian” as a racial category in public health: Implications for communities and practice. American Journal of Public Health 111(11):1969-1975.
Gibson, G. 2008. The environmental contribution to gene expression profiles. Nature Reviews Genetics 9(8):575-581.
Glass, T. A., and M. J. McAtee. 2006. Behavioral science at the crossroads in public health: Extending horizons, envisioning the future. Social Science & Medicine 62(7):1650-1671.
Goicoechea, A. S., F. R. Carnese, C. Dejean, S. A. Avena, T. A. Weimer, M. H. L. P. Franco, S. M. Callegari-Jacques, A. C. Estalote, M. L. Simões, M. Palatnik, and F. M. Salzano. 2001. Genetic relationships between Amerindian populations of Argentina. American Journal of Physical Anthropology 115(2):133-143.
Guardado-Estrada, M., E. Juarez-Torres, I. Medina-Martinez, A. Wegier, A. Macías, G. Gomez, F. Cruz-Talonia, E. Roman-Bassaure, D. Piñero, S. Kofman-Alfaro, and J. Berumen. 2009. A great diversity of Amerindian mitochondrial DNA ancestry is present in the Mexican mestizo population. Journal of Human Genetics 54:695-705.
Guyatt, N. 2016. Bind us apart: How enlightened Americans invented racial segregation. Oxford University Press.
Haak, W., I. Lazaridis, N. Patterson, N. Rohland, S. Mallick, B. Llamas, G. Brandt, S. Nordenfelt, E. Harney, K. Stewardson, Q. Fu, A. Mittnik, E. Bánffy, C. Economou, M. Francken, S. Friederich, R. G. Pena, F. Hallgren, V. Khartanovich, A. Khokhlov, M. Kunst, P. Kuznetsov, H. Meller, O. Mochalov, V. Moiseyev, N. Nicklisch, S. L. Pichler, R. Risch, M. A. Rojo Guerra, C. Roth, A. Szécsényi-Nagy, J. Wahl, M. Meyer, J. Krause, D. Brown, D. Anthony, A. Cooper, K. W. Alt, and D. Reich. 2015. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522(7555):207-211.
Hamilton, T. G. 2019. Immigration and the remaking of black America. New York: Russell Sage Foundation.
Hannaford, I. 1996. Race: The history of an idea in the west. Washington, DC: Woodrow Wilson Center Press.
Hardeman, R. R., P. A. Homan, T. Chantarat, B. A. Davis, and T. H. Brown. 2022. Improving the measurement of structural racism to achieve antiracist health policy. Health Affairs 41(2):179-186.
Hattam, V. 2004. Ethnicity: An American genealogy. In Not just black and white: Historical and contemporary perspectives on immgiration, race, and ethnicity in the United States. Edited by N. Foner and G. M. Fredrickson. New York: Russell Sage Foundation. Pp. 42-60.
Henn, B. M., L. R. Botigué, S. Gravel, W. Wang, A. Brisbin, J. K. Byrnes, K. Fadhlaoui-Zid, P. A. Zalloua, A. Moreno-Estrada, J. Bertranpetit, C. D. Bustamante, and D. Comas. 2012. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genetics 8(1):e1002397.
Hochman, A. 2019. Racialization: A defense of the concept. Ethnic and Racial Studies 42(8):1245-1262.
Homan, P., T. H. Brown, and B. King. 2021. Structural intersectionality as a new direction for health disparities research. Journal of Health and Social Behavior 62(3):350-370.
Hollinger, A. 1998. National culture and communities of descent. Reviews in American History 26(1):312-328.
Hudson, R. R. 1990. Gene genealogies and the coalescent process. Oxford Surveys in Evolutionary Biology 7(1):44.
Jacobson, M. F. 1998. Whiteness of a different color: European immigrants and the alchemy of race. Cambridge, MA: Harvard University Press.
Jelenkovic, A., R. Sund, Y.-M. Hur, Y. Yokoyama, J. v. B. Hjelmborg, S. Möller, C. Honda, P. K. E. Magnusson, N. L. Pedersen, S. Ooki, S. Aaltonen, M. A. Stazi, C. Fagnani, C. D’Ippolito, D. L. Freitas, J. A. Maia, F. Ji, F. Ning, Z. Pang, E. Rebato, A. Busjahn, C. Kandler, K. J. Saudino, K. L. Jang, W. Cozen, A. E. Hwang, T. M. Mack, W. Gao, C. Yu, L. Li, R. P. Corley, B. M. Huibregtse, C. A. Derom, R. F. Vlietinck, R. J. F. Loos, K. Heikkilä, J. Wardle, C. H. Llewellyn, A. Fisher, T. A. McAdams, T. C. Eley, A. M. Gregory, M. He, X. Ding, M. Bjerregaard-Andersen, H. Beck-Nielsen, M. Sodemann, A. D. Tarnoki, D. L. Tarnoki, A. Knafo-Noam, D. Mankuta, L. Abramson, S. A. Burt, K. L. Klump, J. L. Silberg, L. J. Eaves, H. H. Maes, R. F. Krueger, M. McGue, S. Pahlen, M. Gatz, D. A. Butler, M. Bartels, T. C. E. M. van Beijsterveldt, J. M. Craig, R. Saffery, L. Dubois, M. Boivin, M. Brendgen, G. Dionne, F. Vitaro, N. G. Martin, S. E. Medland, G. W. Montgomery, G. E. Swan, R. Krasnow, P. Tynelius, P. Lichtenstein, C. M. A. Haworth, R. Plomin, G. Bayasgalan, D. Narandalai, K. P. Harden, E. M. Tucker-Drob, T. Spector, M. Mangino, G. Lachance, L. A. Baker, C. Tuvblad, G. E. Duncan, D. Buchwald, G. Willemsen, A. Skytthe, K. O. Kyvik, K. Christensen, S. Y. Öncel, F. Aliev, F. Rasmussen, J. H. Goldberg, T. I. A. Sørensen, D. I. Boomsma, J. Kaprio, and K. Silventoinen. 2016. Genetic and environmental influences on height from infancy to early adulthood: An individual-based pooled analysis of 45 twin cohorts. Scientific Reports 6(1):28496.
Kathlene, L., D. Munshi, P. Kurian, and S. L. Morrison. 2022. Cultures in the laboratory: Mapping similarities and differences between Māori and non-Māori in engaging with gene-editing technologies in Aotearoa, New Zealand. Humanities and Social Sciences Communications 9(100).
Katzew, I. 2004. Casta painting: Images of race in eighteenth-century Mexico. New Haven, CT: Yale University Press.
Keel, T. 2018. Divine variations: How Christian thought became racial science. 1st ed. Stanford, CA: Stanford University Press.
Kelleher, J., A. M. Etheridge, and G. McVean. 2016. Efficient coalescent simulation and genealogical analysis for large sample sizes. PLoS Computational Biology 12(5):e1004842.
Koshy, R., A. Ranawat, and V. Scaria. 2017. Al mena: A comprehensive resource of human genetic variants integrating genomes and exomes from Arab, Middle Eastern and north African populations. Journal of Human Genetics 62(10):889-894.
Kowal, E., G. Pearson, C. S. Peacock, S. E. Jamieson, and J. M. Blackwell. 2012. Genetic research and aboriginal and Torres Strait islander Australians. Journal of Bioethical Inquiry 9(4):419-432.
Kramer, M. R., and C. R. Hogue. 2009. Is segregation bad for your health? Epidemiologic Reviews 31(1):178-194.
Krieger, N. 2021. Structural racism, health inequities, and the two-edged sword of data: Structural problems require structural solutions. Frontiers in Public Health 9.
Lee, S. M. 1993. Racial classifications in the U.S. census: 1890–1990. Ethnic and Racial Studies 16(1):75-94.
Lewis, M. W., and K. E. Wigen. 1997. The myth of continents: A critique of metageography. Berkeley, CA: University of California Press.
Li, X., T. Guo, Q. Mu, X. Li, and J. Yu. 2018. Genomic and environmental determinants and their interplay underlying phenotypic plasticity. Proceedings of the National Academy of Sciences 115(26):6679-6684.
Liebler, C. A., S. R. Porter, L. E. Fernandez, J. M. Noon, and S. R. Ennis. 2017. America’s churning races: Race and ethnic response changes between census 2000 and the 2010 census. Demography 54:259-284.
Lipson, M., P.-R. Loh, A. Levin, D. Reich, N. Patterson, and B. Berger. 2013. Efficient moment-based inference of admixture parameters and sources of gene flow. Molecular Biology and Evolution 30(8):1788-1802.
Lucas-Sánchez, M., J. M. Serradell, and D. Comas. 2021. Population history of north Africa based on modern and ancient genomes. Human Molecular Genetics 30(R1):R17-R23.
Mahmud, T. 1999. Colonialism and modern constructions of race: A preliminary inquiry. University of Miami Law Review 53:1219-1249.
Malaspinas, A. S., M. C. Westaway, C. Muller, V. C. Sousa, O. Lao, I. Alves, A. Bergström, G. Athanasiadis, J. Y. Cheng, J. E. Crawford, T. H. Heupink, E. Macholdt, S. Peischl, S. Rasmussen, S. Schiffels, S. Subramanian, J. L. Wright, A. Albrechtsen, C. Barbieri, I. Dupanloup, A. Eriksson, A. Margaryan, I. Moltke, I. Pugach, T. S. Korneliussen, I. P. Levkivskyi, J. V. Moreno-Mayar, S. Ni, F. Racimo, M. Sikora, Y. Xue, F. A. Aghakhanian, N. Brucato, S. Brunak, P. F. Campos, W. Clark, S. Ellingvåg, G. Fourmile, P. Gerbault, D. Injie, G. Koki, M. Leavesley, B. Logan, A. Lynch, E. A. Matisoo-Smith, P. J. McAllister, A. J. Mentzer, M. Metspalu, A. B. Migliano, L. Murgha, M. E. Phipps, W. Pomat, D. Reynolds, F. X. Ricaut, P. Siba, M. G. Thomas, T. Wales, C. M. Wall, S. J. Oppenheimer, C. Tyler-Smith, R. Durbin, J. Dortch, A. Manica, M. H. Schierup, R. A. Foley, M. M. Lahr, C. Bowern, J. D. Wall, T. Mailund, M. Stoneking, R. Nielsen, M. S. Sandhu, L. Excoffier, D. M. Lambert, and E. Willerslev. 2016. A genomic history of aboriginal Australia. Nature 538(7624):207-214.
Malinowska, J., and T. Żuradzki. 2022. Reductionist methodology and the ambiguity of the categories of race and ethnicity in biomedical research: An exploratory study of recent evidence. Medicine, Health Care and Philosophy 26(3):1-14.
Marca-Ysabel, M. V., F. Rajabli, M. Cornejo-Olivas, P. G. Whitehead, N. K. Hofmann, M. Z. Illanes Manrique, D. M. Veliz Otani, A. K. Milla Neyra, S. Castro Suarez, M. Meza Vega, L. D. Adams, P. R. Mena, I. Rosario, M. L. Cuccaro, J. M. Vance, G. W. Beecham, N. Custodio, R. Montesinos, P. E. Mazzetti Soler, and M. A. Pericak-Vance. 2021. Dissecting the role of Amerindian genetic ancestry and the ApoE ε4 allele on Alzheimer disease in an admixed Peruvian population. Neurobiology and Aging 101:298.e211-298.e215.
Martin, C. L., L. Ghastine, E. K. Lodge, R. Dhingra, and C. K. Ward-Caviness. 2022. Understanding health inequalities through the lens of social epigenetics. Annual Review of Public Health 43:235-254.
Martinez, R. A. M., N. Andrabi, A. N. Goodwin, R. E. Wilbur, N. R. Smith, and P. N. Zivich. 2022. Conceptualization, operationalization, and utilization of race and ethnicity in major epidemiology journals, 1995–2018: A systematic review. American Journal of Epidemiology 192(3):483-496.
Martínez Cobo, J. R. 1987. Study of the problem of discrimination against indigenous populations: Conclusions, proposals and recommendation. Vol. V. New York: United Nations.
Mathieson, I., and A. Scally. 2020. What is ancestry? PLoS Genetics 16(3):e1008624.
Mayhew, A. J., and D. Meyre. 2017. Assessing the heritability of complex traits in humans: Methodological challenges and opportunities. Current Genomics 18(4):332-340.
Monk, E. P. 2022. Inequality without groups: Contemporary theories of categories, intersectional typicality, and the disaggregation of difference. Sociological Theory 40(1):3–27.
Moreno Navarro, I. 1973. Los “cuadros del mestizaje americano”: Estudio antropológico del mestizaje. Madrid, Spain: Ediciones Jose Porrua Turanzas.
Morgan, E. S. 1975. American slavery, American freedom: The ordeal of colonial Virginia. New York: W.W. Norton.
Morning, A. 2008. Ethnic classification in global perspective: A cross-national survey of the 2000 census round. Population Research and Policy Review 27:239-272.
Morning, A., and M. Maneri. 2022. An ugly word: Rethinking race in Italy and the United States. New York: Russell Sage Foundation.
Nasir, B. F., R. Vinayagam, and K. Rae. 2022. “It’s what makes us unique”: Indigenous Australian perspectives on genetics research to improve comorbid mental and chronic disease outcomes. Current Medical Research and Opinion 38(7):1219-1228.
NCAI (National Congress of American Indians). 2020. Tribal nations and the United States: An introduction. Washington, DC: National Congress of American Indians.
NRC (National Research Council). 1991. The children of atomic bomb survivors: A genetic study. Edited by J. V. Neel and W. J. Schull. Washington, DC: National Academy Press.
Parks, T. 2016. How racism, segregation drive health disparities. In Patient support & advocacy. Chicago, IL: American Medical Association.
Patterson, N., P. Moorjani, Y. Luo, S. Mallick, N. Rohland, Y. Zhan, T. Genschoreck, T. Webster, and D. Reich. 2012. Ancient admixture in human history. Genetics 192(3):1065-1093.
Penner, A. M., and A. Saperstein. 2008. How social status shapes race. Proceedings of the National Academy of Sciences 105:19628-19630.
Peters, E. J. 2011. Still invisible: Enumeration of indigenous peoples in census questionnaires internationally. Aboriginal Policy Studies 1(2):68-100.
Pickrell, J. K., and J. K. Pritchard. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genetics 8(11):e1002967.
Prewitt, K. 2005. Racial classification in America: Where do we go from here? Daedalus 134(1):5-17.
Pritchard, J. K., M. Stephens, and P. Donnelly. 2000. Inference of population structure using multilocus genotype data. Genetics 155(2):945-959.
Rasmussen, M., X. Guo, Y. Wang, K. E. Lohmueller, S. Rasmussen, A. Albrechtsen, L. Skotte, S. Lindgreen, M. Metspalu, T. Jombart, T. Kivisild, W. Zhai, A. Eriksson, A. Manica, L. Orlando, F. M. De La Vega, S. Tridico, E. Metspalu, K. Nielsen, M. C. Ávila-Arcos, J. V. Moreno-Mayar, C. Muller, J. Dortch, M. T. Gilbert, O. Lund, A. Wesolowska, M. Karmin, L. A. Weinert, B. Wang, J. Li, S. Tai, F. Xiao, T. Hanihara, G. van Driem, A. R. Jha, F. X. Ricaut, P. de Knijff, A. B. Migliano, I. Gallego Romero, K. Kristiansen, D. M. Lambert, S. Brunak, P. Forster, B. Brinkmann, O. Nehlich, M. Bunce, M. Richards, R. Gupta, C. D. Bustamante, A. Krogh, R. A. Foley, M. M. Lahr, F. Balloux, T. Sicheritz-Pontén, R. Villems, R. Nielsen, J. Wang, and E. Willerslev. 2011. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science 334(6052):94-98.
Reich, D. 2018. Who we are and how we got here: Ancient DNA and the new science of the human past. Oxford University Press.
Reich, D., A. L. Price, and N. Patterson. 2008. Principal component analysis of genetic data. Nature Genetics 40(5):491-492.
Roberts, D. 2011. Fatal invention: How science, politics, and big business re-create race in the twenty-first century. New York: The New Press.
Robette, N. 2022. The dead ends of sociogenomics. Population 77(2):181-216.
Rodriguez-Flores, J. L., K. Fakhro, F. Agosto-Perez, M. D. Ramstetter, L. Arbiza, T. L. Vincent, A. Robay, J. A. Malek, K. Suhre, L. Chouchane, R. Badii, A. Al-Nabet Al-Marri, C. Abi Khalil, M. Zirie, A. Jayyousi, J. Salit, A. Keinan, A. G. Clark, R. G. Crystal, and J. G. Mezey. 2016. Indigenous Arabs are descendants of the earliest split from ancient Eurasian populations. Genome Research 26(2):151-162.
Rodriguez Mega, E. 2021. How the mixed-race mestizo myth warped science in Latin America. Nature 600(7889):374-378.
Rohde, D. L. T., S. Olson, and J. T. Chang. 2004. Modelling the recent common ancestry of all living humans. Nature 431(7008):562-566.
Rosenberg, N. A. 2021. A population-genetic perspective on the similarities and differences among worldwide human populations. Human Biology 92(3):135-152.
Rosenberg, N. A., J. K. Pritchard, J. L. Weber, H. M. Cann, K. K. Kidd, L. A. Zhivotovsky, and M. W. Feldman. 2002. Genetic structure of human populations. Science 298(5602):2381-2385.
Roth, W. D. 2016. The multiple dimensions of race. Ethnic and Racial Studies 39:1310-1338.
Saperstein, A., and A. M. Penner. 2012. Racial fluidity and inequality in the United States. American Journal of Sociology 118:676-727.
Schaefer, N. K., B. Shapiro, and R. E. Green. 2021. An ancestral recombination graph of human, Neanderthal, and Denisovan genomes. Scientific Advances 7(29):eabc0776.
Seabrook, J. A., and W. R. Avison. 2010. Genotype–environment interaction and sociology: Contributions and complexities. Social Science & Medicine 70(9):1277-1284.
Shanahan, M. J., and S. M. Hofer. 2011. Molecular genetics, aging, and well-being: Sensitive period, accumulation, and pathway models. In Handbook of aging and the social sciences. 7th ed., edited by L. K. George. Elsevier. Pp. 135-147.
Silva-Zolezzi, I., A. Hidalgo-Miranda, J. Estrada-Gil, J. C. Fernandez-Lopez, L. Uribe-Figueroa, A. Contreras, E. Balam-Ortiz, L. del Bosque-Plata, D. Velazquez-Fernandez, C. Lara, R. Goya, E. Hernandez-Lemus, C. Davila, E. Barrientos, S. March, and G. Jimenez-Sanchez. 2009. Analysis of genomic diversity in Mexican mestizo populations to develop genomic medicine in Mexico. Proceedings of the National Academy of Sciences 106(21):8611-8616.
Smedley, A., and B. D. Smedley. 2012. Race in North America: Origin and evolution of a worldview. 4th ed. Oxfordshire, UK: Routledge.
Smith, E. N., and L. Kruglyak. 2008. Gene–environment interaction in yeast gene expression. PLoS Biology 6(4):e83.
Snipp, C. M. 2002. American Indians: Clues to the future of other racial groups. In The new race question: How the census counts multiracial individuals, edited by J. Perlmann and M. C. Waters. New York and Annandale-on-Hudson, NY: Russell Sage Foundation and Levy Economics Institute of Bard College. Pp. 189-214.
Snipp, C. M., and National Committee for Research on the 1980 Census. 1989. American Indians: The first of this land. New York: Russell Sage Foundation.
Spangenberg, L., M. I. Fariello, D. Arce, G. Illanes, G. Greif, J.-Y. Shin, S. K. Yoo, J. S. Seo, C. Robello, C. Kim, J. Novembre, M. Sans, and H. Naya. 2021. Indigenous ancestry and admixture in the Uruguayan population. Frontiers of Genetics 12:733195.
Sparrow, D. B., G. Chapman, A. J. Smith, M. Z. Mattar, J. Major, V. C. O’Reilly, Y. Saga, E. Zackai, J. Dormans, A. Benjamin, L. McGregor, R. Kageyama, K. Kusumi, and S. Dunmoodie. 2012. A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell 149(2):295-306.
Spruhan, P. 2017. CDIB: The role of the certificate of degree of Indian blood in defining Native American legal identity. American Indian Law Journal 6:169.
Strickberger, M. W. 1985. Genetics. 3rd ed. New York: Macmillan.
The Swedish Development Forum. 2018. Europe’s only recognized indigenous peoples live in Sweden. https://fuf.se/en/magasin/europas-enda-erkanda-urfolk-bor-i-sverigre/ (accessed December 13, 2022).
Tätte, K., E. Metspalu, H. Post, L. Palencia-Madrid, J. R. Luis, M. Reidla, E. Tamm, A. M. Ilumäe, M. M. de Pancorbo, R. Garcia-Bertrand, M. Metspalu, and R. J. Herrera. 2022. Genetic characterization of populations in the Marquesas Archipelago in the context of the Austronesian expansion. Scientific Reports 12:5312.
Thompson, E. A. 2013. Identity by descent: Variation in meiosis, across genomes, and in populations. Genetics 194(2):301-326.
Tishkoff, S. A., F. A. Reed, F. R. Friedlaender, C. Ehret, A. Ranciaro, A. Froment, J. B. Hirbo, A. A. Awomoyi, J. M. Bodo, O. Doumbo, M. Ibrahim, A. T. Juma, M. J. Kotze, G. Lema, J. H. Moore, H. Mortensen, T. B. Nyambo, S. A. Omar, K. Powell, G. S. Pretorius, M. W. Smith, M. A. Thera, C. Wambebe, J. L. Weber, and S. M. Williams. 2009. The genetic structure and history of Africans and African Americans. Science 324(5930):1035-1044.
Umaefulam, V., T. Kleissen, and C. Barnabe. 2022. The representation of indigenous peoples in chronic disease clinical trials in Australia, Canada, New Zealand, and the United States. Clinical Trials 19(1):22-32.
U.S. Department of the Interior. n.d. Tribal enrollment process. https://www.doi.gov/tribes/enrollment#main-content (accessed December 14, 2022).
Vargas, N., and J. Kingsbury. 2016. Racial identity contestation: Mapping and measuring racial boundaries. Sociology Compass 10(8):718-729.
Wang, K., M. Cadzow, M. Bixley, M. P. Leask, M. E. Merriman, Q. Yang, Z. Li, R. Takei, A. Phipps-Green, T. J. Major, R. Topless, N. Dalbeth, F. King, R. Murphy, L. K. Stamp, J. Zoysa, Z. Wang, Y. Shi, and T. R. Merriman. 2022. A Polynesian-specific copy number variant encompassing the MICA gene associates with gout. Human Molecular Genetics 31(21):3757-3768.
Wang, S., C. M. Lewis, Jr., M. Jakobsson, S. Ramachandran, N. Ray, G. Bedoya, W. Rojas, M. V. Parra, J. A. Molina, C. Gallo, G. Mazzotti, G. Poletti, K. Hill, A. M. Hurtado, D. Labuda, W. Klitz, R. Barrantes, M. C. Bortolini, F. M. Salzano, M. L. Petzl-Erler, L. T. Tsuneto, E. Llop, F. Rothhammer, L. Excoffier, M. W. Feldman, N. A. Rosenberg, and A. Ruiz-Linares. 2007. Genetic variation and population structure in Native Americans. PLoS Genetics 3(11):e185.
Wang, S., N. Ray, W. Rojas, M. V. Parra, G. Bedoya, C. Gallo, G. Poletti, G. Mazzotti, K. Hill, A. M. Hurtado, B. Camrena, H. Nicolini, W. Klitz, R. Barrantes, J. A. Molina, N. B. Freimer, M. C. Bortolini, F. M. Salzano, M. L. Petzl-Erler, L. T. Tsuneto, J. E. Dipierri, E. L. Alfaro, G. Bailliet, N. O. Bianchi, E. Llop, F. Rothhammer, L. Excoffier, and A. Ruiz-Linares. 2008. Geographic patterns of genome admixture in Latin American mestizos. PLoS Genetics 4(3):e1000037.
Waters, M. C. 1990. Ethnic options: Choosing identities in America. Berkeley, CA: University of California Press.
Weyrich, A., D. Lenz, and J. Fickel. 2018. Environmental change-dependent inherited epigenetic response. Genes 10(4).
Williams, D. R., and C. Collins. 2001. Racial residential segregation: A fundamental cause of racial disparities in health. Public Health Reports 116(5):404-416.
Williams, D. R., J. A. Lawrence, and B. A. Davis. 2019. Racism and health: Evidence and needed research. Annual Review of Public Health 40:105-125.
Wimmer, A. 2013. Ethnic boundary making: Institutions, power, networks. Oxford, UK: Oxford University Press.
Wiuf, C., and J. Hein. 1999. Recombination as a point process along sequences. Theoretical Population Biology 55(3):248-259.
Wohns, A. W., Y. Wong, B. Jeffery, A. Akbari, S. Mallick, R. Pinhasi, N. Patterson, D. Reich, J. Kelleher, and G. McVean. 2022. A unified genealogy of modern and ancient genomes. Science 375(6583):eabi8264.
Wolfe, P. 2001. Land, labor, and difference: Elementary structures of race. American Historical Review 106(3):866-905.
Wolfe, P. 2016. Traces of history: Elementary structures of race. London: Verso Books.


































