4
Crosscutting Issues in Autoimmune Diseases
In Chapter 1 and Chapter 3, the committee highlighted unique and heterogeneous features of specific autoimmune diseases. In contrast, this chapter examines common features found among many autoimmune diseases. Because discussion applies to autoimmune diseases as a category of diseases, the 11 diseases of focus as well as other autoimmune diseases, such as alopecia areata, juvenile idiopathic arthritis, myocarditis, spondyloarthropathy, systemic sclerosis (also known as scleroderma), and vasculitis, are referenced.
COMMONALITIES ACROSS AUTOIMMUNE DISEASES
While every autoimmune disease has its unique characteristics, many of them also share common features that researchers believe—and evidence supports—suggest that there are common mechanisms involved in the etiology of autoimmune diseases. For example, the fact that autoimmune diseases tend to occur in family clusters suggests that there are common genetic and environmental factors involved in triggering autoimmunity and disease symptoms. Research shows that genetic factors alone cannot explain the etiology of any autoimmune disease, and that environmental agents or insults—infectious microorganisms and respiratory and dietary exposures are leading candidates for several autoimmune diseases—are interacting with those genetic factors to produce the disease state. Certain types of proinflammatory immune responses and the presence of autoantibodies are two other common features. Finally,
multiple autoimmune diseases show sex differences in disease incidence and severity. The discussion below highlights how these commonalities, as well as common mechanistic pathways, can serve as therapeutic targets.
Genetics
Research suggests strongly that autoimmunity leading to autoimmune disease requires at least one, but likely more than one, gene (Sogkas et al., 2021) that interacts with environmental factors. The gene–environment relationship is complex, with environmental changes altering genes through epigenetic modifications that not only affect that individual but subsequent offspring (Cavalli and Heard, 2019; Surace and Hedrich, 2019). Epigenetic factors could contribute to early onset of some autoimmune diseases such juvenile idiopathic arthritis (Meyer et al., 2015). Individuals with one autoimmune disease are more likely to develop another autoimmune disease (Cojocaru et al., 2010), and autoimmune diseases, although often not the same autoimmune disease, typically cluster in families (Shamim and Miller, 2000). In addition, autoantibodies are more likely to develop in family members, whether symptomatic or not, of an individual with an autoimmune disease (James et al., 2019; Leslie et al., 2001). These findings, along with the observation that autoimmune diseases run in families, lead the research community to believe that common mechanisms increase autoimmunity in genetically susceptible individuals.
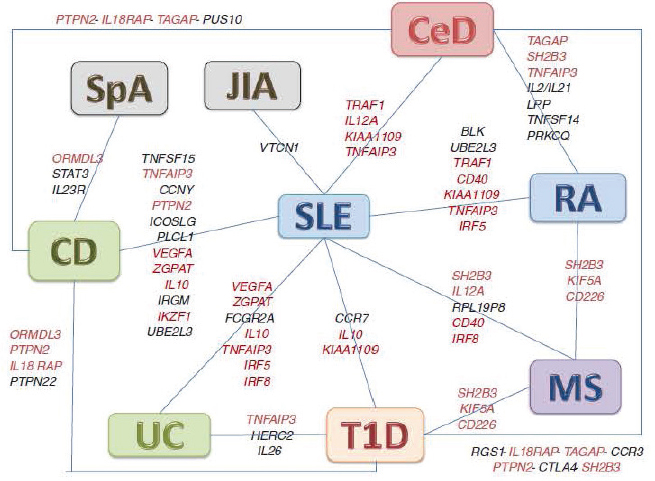
Human leukocyte antigen (HLA), or HLA haplotype, is the best available predictor of developing an autoimmune disease (Bacon et al., 1974; Cooper et al., 1999; Matzaraki et al., 2017) and is directly related to increased autoantibodies in family members (Wong and Wen, 2003), though it does not appear to be the only region of the genome where different autoimmune diseases have genes in common (Cho and Gregersen, 2011; Cooper et al., 1999). Many of the genes that confer susceptibility to autoimmune disease and that different autoimmune diseases share regulate the immune response (Sogkas et al., 2021). Some autoimmune diseases with different clinical characteristics, including which organs are affected, share genetic variants, indicating that these diseases may share some common pathways of immune dysfunction (see Figure 4-1 and Figure 4-2). This supports the suggestion that the specific organs targeted in individuals with similar genetic risk variants, such as in families with multiple affected individuals, may require an additional factor such as an environmental exposure for disease to progress. In addition to the 11 diseases of focus, Figure 4-1 and Figure 4-2 include systemic sclerosis, spondyloarthropathy, primary sclerosing cholangitis, and juvenile

AS, ankylosing spondylitis; ATD, autoimmune thyroid disease; IBD, inflammatory bowel disease (includes Crohn’s disease and ulcerative colitis); MS, multiple sclerosis; PBC, primary biliary cholangitis/cirrhosis; PSC, primary sclerosing cholangitis; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; SSc, systemic sclerosis; T1D, type 1 diabetes.
SOURCES: Acosta-Herrera et al., 2019; Anderson et al., 2011; Andlauer et al., 2016; Barrett et al., 2009; Barrett et al., 2008; Bentham et al., 2015; de Lange et al., 2017; Dubois et al., 2010; Ellinghaus et al., 2016; Eyre et al., 2012; Gulamhusein et al., 2015; Imgenberg-Kreuz et al., 2021; International Genetics of Ankylosing Spondylitis Consortium (IGAS) et al., 2013; International Multiple Sclerosis Genetics Consortium (IMSGC), 2019; International Multiple Sclerosis Genetics Consortium (IMSGC) et al., 2013; Jostins et al., 2012; Khatri et al., 2020; Kochi, 2016; Langefeld et al., 2017; Lessard et al., 2016; Li et al., 2015; Márquez et al., 2018; Morris et al., 2016; Okada et al., 2014; Onengut-Gumuscu et al., 2015; Qiu et al., 2017; Radstake et al., 2010; Saevarsdottir et al., 2020; Sawcer et al., 2011; Stahl et al., 2010; Todd et al., 2007; Tsoi et al., 2012, 2017; Wang et al., 2018, 2021; Wellcome Trust Case Control Consortium et al., 2007.
idiopathic arthritis, further highlighting commonalities among autoimmune diseases.
Determining genetic risk for autoimmune disease is complex because many different genes may be associated with the risk for the disease. Polygenic risk scores are used to convert the complexity of the multiple genetic risk variants into a single quantified measure (NHGRI, 2020). Studies defining polygenic risk scores for specific autoimmune diseases have demonstrated potential utility in diagnosis, prognosis, and molecular subsetting of autoimmune diseases. Recent studies in systemic lupus erythematosus (SLE) found associations of increased organ damage and mortality in individuals with high genetic risk scores (Reid et al., 2020). Another study focused on genes in different immunological pathways in order to identify different molecular subsets of individuals with SLE based on pathway-specific genetic risk scores (Sandling et al., 2021). Genetic risk scores have been used in studies of other autoimmune diseases such as type 1 diabetes (Sharp et al., 2018), multiple sclerosis (de Mol et al., 2020; van Pelt et al., 2013), and systemic sclerosis (Bossini-Castillo et al., 2021). In inflammatory bowel disease (IBD), genetic risk scoring is less well-established but shows some promise (Abakkouy and Cleynen, 2021). Advantages of genetic risk scores include the ease of a standardized measurement and the absence of disease activity-based variability that can complicate measures of other disease-associated biomarkers (Sharp et al., 2018). Disadvantages include risk scores that represent relative risk rather than absolute risk, and that their applicability to the general population will require studying more diverse populations. Much of the available genomics data are based on individuals of European ancestry, which makes the relevance of these risk scores less clear for other populations (NHGRI, 2020). Moreover, the overlap in genetic risk between autoimmune diseases such as systemic sclerosis and SLE, Sjögren’s disease, or rheumatoid arthritis may preclude the ability to accurately distinguish the diseases based on genetic risk scores alone (Bossini-Castillo et al., 2021). Thus, additional studies are needed to overcome the limitations of polygenic risk scores before they can be used clinically, but such studies are warranted to help shift the diagnosis and treatment of autoimmune diseases to prediction and prevention.
Despite clear associations between genetic variants of immune-related genes and specific autoimmune diseases, a complete understanding of the specific effects of these variants in the pathogenesis of these diseases is lacking. The altered biological functions of the genetic variants and their roles in immune dysregulation remain gaps in our knowledge of the genotype–phenotype relationships of disease-associated genetic variants. An important focus of future research will be to systematically map the biologic function of genetic variants for the different autoimmune diseases. Defining the functions of variant-related immune pathways is
critical as this knowledge can inform clinical approaches to modulate the dysregulated or overactive immune pathways. The same pathway may or may not function the same way across diseases. For example, the pathogenic role of tumor necrosis factor (TNF) in rheumatoid arthritis, inflammatory bowel disease, and psoriasis led to targeting therapies to block TNF to treat these diseases (Jang et al., 2021), but TNF inhibitors have negative effects in the treatment of multiple sclerosis (Kemanetzoglou and Andreadou, 2017). In some individuals without multiple sclerosis, treatment with TNF inhibitors can cause multiple sclerosis-like demyelination (Kemanetzoglou and Andreadou, 2017). Despite effectively treating psoriasis in many individuals, the use of TNF inhibitors may also induce psoriasis (Mazloom et al., 2020). Similarly, genetic variants may have opposite effects in different autoimmune diseases and be associated with protective effects in some autoimmune diseases but with increased risk in others (Dendrou et al., 2016; Wang et al., 2010). Beyond associations of gene
NOTES: There are more recent data, and the data continually grow. This figure is not intended to be comprehensive.
CD, Crohn’s disease; CeD, celiac disease; JIA, juvenile idiopathic arthritis; MS, multiple sclerosis; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; SpA, spondyloarthropathy; T1D, type 1 diabetes; UC, ulcerative colitis.
SOURCE: Richard-Miceli and Criswell, 2012.
variants with autoimmune diseases, altered expression of genes through epigenetic modifications may drive differential effects of immune-related genes (Mazzone et al., 2019).
Finding: Studies of genetics in autoimmune diseases have identified genes associated with specific autoimmune diseases. The overlap in genes associated with different autoimmune diseases underscores the contribution of non-genetic factors, such as environmental exposures, in determining which organ systems are targeted by the autoimmune response, but the mechanisms of the genotype–phenotype associations are not known.
Finding: Studies have consolidated the complex gene variants into a single genetic risk score to define genetic risk for a specific autoimmune disease. However, the majority of available data driving genetic risk scores are based on individuals of European ancestry.
Conclusion: Genetic risks are common and often overlapping in autoimmune diseases, but more research is needed to define these risks in different populations in order to develop more widely applicable genetic risk scores and to promote studies to define genotype-phenotype associations.
Environmental Factors
Studies of the incidence of autoimmune disease in identical and fraternal twins show that autoimmune diseases may require an environmental signal or exposure to develop (Figure 4-3). Nationwide or population-based twin registry or cohort studies comparing the incidence of SLE (Ulff-Møller et al., 2018), type 1 diabetes (Nistico et al., 2012), multiple sclerosis (Willer et al., 2003), and Graves’ disease (Brix et al., 2001) find probandwise concordance rates of 25 to 45 percent in identical twins and much lower rates, ranging from 5 to 16 percent, in fraternal twins, implying strong genetic factors and the need for a “second hit”—presumably environmental exposure—for the development of autoimmune disease. Investigators found a lower concordance rate of 9.1 percent in identical twins in rheumatoid arthritis, similar to the 6.4 percent concordance rate seen in fraternal twins (Svendsen et al., 2013). In systemic sclerosis, identical and fraternal twins both had a 5 percent concordance rate (Feghali-Bostwick et al., 2003), suggesting more environmental and less genetic influence on disease development.
Because they depend on diagnosis definition and duration of observation, concordance rates may mislead. For example, in a restudy that applied new technology to examine twin concordance rates fifteen years
after an earlier study, the researchers showed that SLE markers had been present in the well twins of the earlier study (Reichlin et al., 1992), and thus the well twins had been serologically but not clinically concordant with their siblings. The restudy also showed that some well twins developed clinical illness, becoming clinically concordant, decades after their twin siblings were first diagnosed. These observations strongly support the “second-hit” hypothesis, which holds that a genetic profile requires an additional exposure event to become clinical illness. Examples of exposures associated with autoimmune diseases include infectious agents, pesticides, solvents, endocrine-disrupting chemicals, occupational exposure to respirable particulates and fibers, and personal factors such as cigarette-smoking history and diet preferences (Cooper et al., 2009; Cunningham, 2019; Diegel et al., 2018; Hahn et al., 2022; Khan and Wang, 2019; Parks et al., 1999; Popescu et al., 2021).
Infections
For some autoimmune diseases, such as rheumatic fever, myocarditis, multiple sclerosis, rheumatoid arthritis, and SLE, research has identified a causal link between bacterial or viral infection and the development of disease (Bjornevik et al., 2022; Cunningham, 2019; Fairweather et al., 2001, 2003; Fechtner et al., 2021; Jog et al., 2019; Lanz et al., 2022; McClain et al., 2005). For example, data have found that infection with or reactivation of Epstein-Barr virus precedes development of multiple sclerosis, rheumatoid arthritis, and SLE (Bjornevik et al., 2022; Fechtner et al., 2021; Jog et al., 2019; Lanz et al., 2022; McClain et al., 2005).
Additionally, research has found associations between type 1 diabetes and coxsackievirus and cytomegalovirus infections (Fairweather et al., 2001; Rodriguez-Calvo, 2019); multiple sclerosis and Epstein-Barr virus (Bjornevik et al., 2022) and measles infections (Mentis et al., 2017; Persson et al., 2014; Tucker and Andrew Paskauskas, 2008); and rheumatoid arthritis and mycobacteria and Epstein-Barr virus infections (Fairweather et al., 2012). However, most of the associations between infectious agents and autoimmune diseases are considered to be circumstantial. Linking an infection to an autoimmune disease is problematic because there is usually an extended period between the infection and the appearance of clinical autoimmune disease. The best evidence that infectious agents can cause autoimmune disease comes from animal studies in which researchers administered self-antigens with adjuvant incorporating non-infectious microbial antigens (Fairweather et al., 2001). A variation on the classic experimental autoimmune model uses an injection of heart- or salivary gland-derived cytomegalovirus or coxsackievirus B3 as the trigger to
induce myocarditis, which is associated with the development of autoantibodies against cardiac myosin (Fairweather et al., 2001).
Some of the best clinical evidence that viral infections can cause an autoimmune disease comes from the ability of SARS-CoV-2 to cause myocarditis, which occurs in approximately 4.5 percent of COVID-19 cases (Kawakami et al., 2021). SARS-CoV-2 has also been associated with the production of multiple autoantibodies and the development of autoimmune diseases besides myocarditis such as antiphospholipid syndrome, Guillain-Barré syndrome, and Kawasaki disease (Dotan et al., 2021). The production of autoantibodies after viral infections is a feature of animal models of virally induced autoimmune disease (Fairweather et al., 2001), further indicating this as an important area for future investigation. Multiple diverse microorganisms have been associated with a single autoimmune disease—for example, several different viral infections are associated with myocarditis—which suggests that infectious agents induce autoimmune disease through common mechanisms (Fairweather and Rose, 2006). Similarly, infection by one microorganism may induce different autoimmune diseases. Coxsackievirus is associated with type 1 diabetes (Rodriguez-Calvo, 2019), autoimmune thyroiditis (Hashimoto’s thyroiditis) (Mori and Yoshida, 2010), and myocarditis (Tam, 2006), and in children, SARS-CoV-2 infection is linked to myocarditis (Boehmer et al., 2021) and Kawasaki-like inflammatory disease (Dotan et al., 2021) as well as multisystem inflammatory syndrome (Consiglio et al., 2020). The inflammatory response to infection may play a greater role in causing autoimmunity than the specific infectious agent; the inflammation activates common pathogenic mechanisms that cause autoimmune disease. Inherent in this observation is that common innate immune factors such as Toll-like receptor 2 (TLR2) and/or TLR4 are able to initiate the autoimmune response to infectious agents (Mohammad Hosseini et al., 2015).
There are a number of theories for how infections or other environmental agents can trigger autoimmunity and autoimmune disease. These include (Cavalli and Heard, 2019; Fairweather and Rose, 2006; Surace and Hedrich, 2019):
- direct viral damage;
- release of cryptic self-peptides, which are antigens that are normally exposed to the immune system in such low amounts that the body has not developed self-tolerance1 to them;
___________________
1 Self-tolerance refers to the immune system’s tolerance to the body’s own antigens (self-antigens). By this mechanism, the immune system prevents the production of functional B cells and T cells that react to self-antigens and prevents the body from directing an immune response against itself. Impairment of this mechanism leads to autoimmunity and production of autoantibodies against self-antigens (Hale et al., 2005).
- antigenic spread, which occurs when the immune system’s response to a microbial antigen expands to include similar self-antigens;
- molecular mimicry, which occurs when a microbial antigen closely resembles a self-antigen;
- epigenetics;
- bystander activation, which occurs when the immune system is activated in an antigen-independent manner and then produces a cytokine storm; and
- an adjuvant effect, which occurs when a microbial antigen or chemical adjuvant activates the innate immune response to self-tissue. Support for the latter comes from animal experiments in which researchers induce autoimmune disease using self-peptides and adjuvants; examples include inducing rheumatoid arthritis by administering collagen with an adjuvant, multiple sclerosis by inoculating with myelin basic protein with an adjuvant, and myocarditis by injecting cardiac myosin with an adjuvant.
Other hypotheses hold that co-infections with bacteria and viruses are necessary to induce autoimmune disease or that autoimmunity may occur commonly as a normal response needed to clear damaged self (Root-Bernstein and Fairweather, 2015).
Respiratory Exposures: Silica, Asbestos, and Tobacco Smoke
The research pertaining to respirable silica exposure and systemic autoimmune diseases—rheumatoid arthritis, SLE, systemic sclerosis, and various forms of small-vessel vasculitis—spans more than 100 years (Parks et al., 1999). A robust set of studies, with highly consistent findings from a variety of populations and settings and using different study designs, have demonstrated at least a 2- to 4-fold increased risk of developing each of those diseases associated with occupational exposure to silica (Figure 4-4). The figure’s inclusion of data on 2 of the 11 diseases of focus as well as systemic sclerosis and vasculitis highlights the risk silica exposure carries across autoimmune diseases.
Studies have also investigated immune-related effects of silica in the lung. Silica stimulates macrophages and also leads to their destruction, releasing intracellular self-antigens and stimulating proinflammatory cytokines, including interleukin 1 (IL-1) and TNF, and eventually activating the innate and adaptive immune response in the lungs and other tissues (Bates et al., 2015; Brown et al., 2003; Chauhan et al., 2021; Cooper et al., 2008; Foster et al., 2019). Studies of a breed of mice susceptible to SLE have examined the mechanisms by which inhaled silica induces both
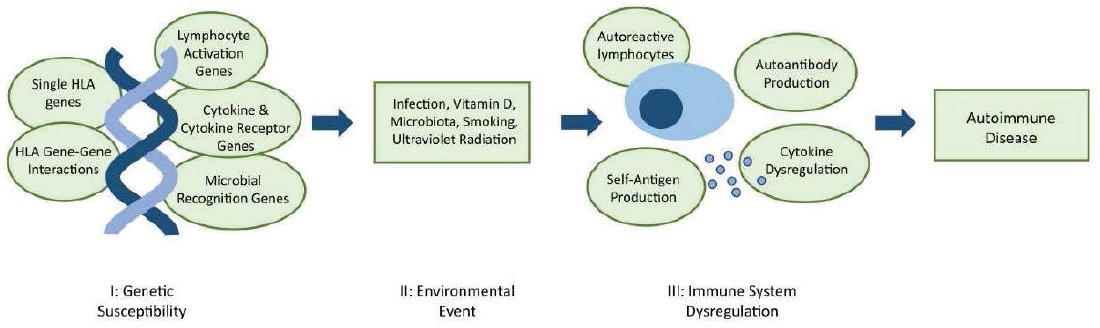
NOTE: HLA, human leukocyte antigen.
SOURCE: Reprinted with permission from Stafford et al., 2020.
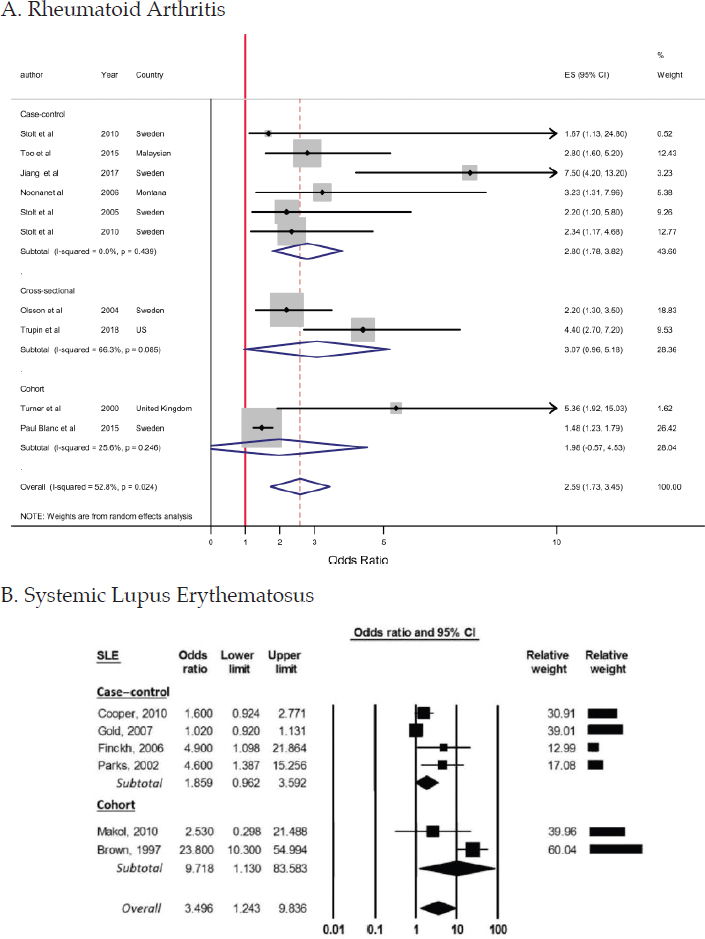
NOTES: More recent studies in each of these diseases provide additional support for this association (Boudigaard et al., 2021; De Decker et al., 2018; Giorgiutti et al., 2021; Ilar et al., 2019).
ANCA, antineutrophil cytoplasmic antibody; CI, confidence interval; ES, effect size; OR, odds ratio.
SOURCES: A. reproduced from Mehri et al., 2020. B. reproduced from Morotti et al., 2021. C. reproduced from Gómez-Puerta et al., 2013. D. reproduced from Rubio-Rivas et al., 2017.
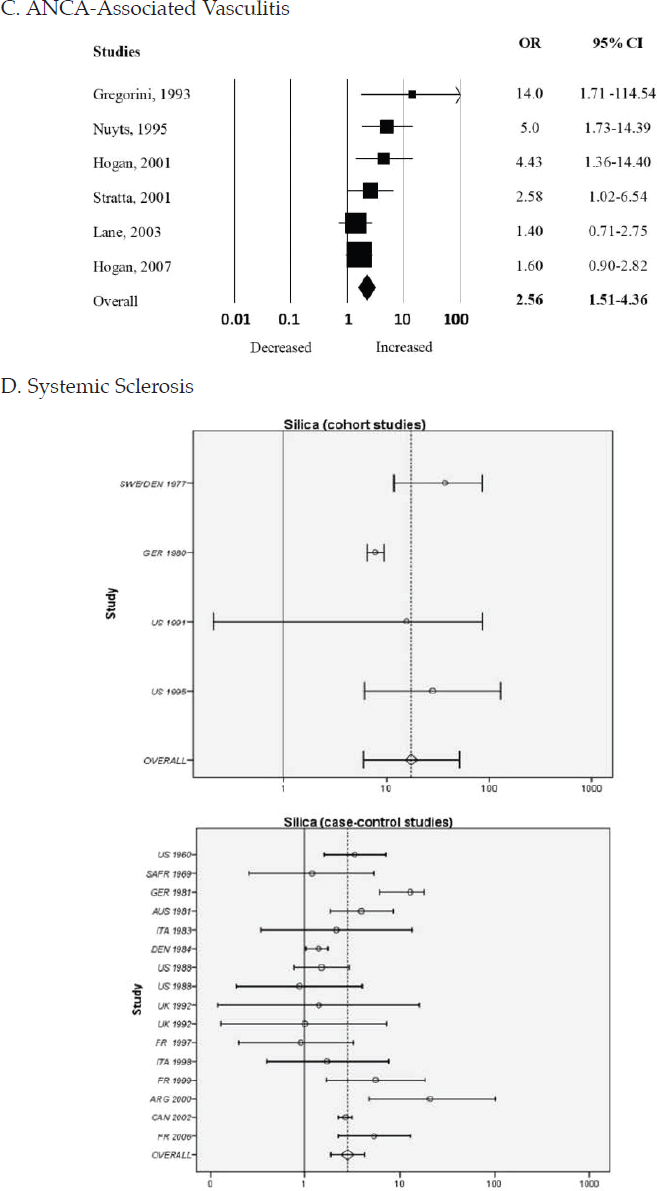
development and exacerbation of SLE and activates immune responses outside the lung (Bates et al., 2015; Brown et al., 2003; Chauhan et al., 2021; Foster et al., 2019). Examining silica in combination with other exposures has also been revealing. A recent study using a breed of mice that is not prone to autoimmune disease found that an early-life exposure to lymphocytic choriomeningitis virus followed later by a single high-intensity airway exposure to silica in the adult mice produced lupus-related autoantibodies and kidney lesions (Gonzalez-Quintial et al., 2019).
Recent research has expanded the interest in respirable exposures to other particles and fibers. Several studies have examined systemic autoimmune diseases and autoantibodies in populations exposed to various forms of asbestos and asbestiform particles in Montana and in Australia, Italy, Korea, and Sweden (Carey et al., 2021; Diegel et al., 2018; Ilar et al., 2019; Lee et al., 2020; Noonan et al., 2006; Pfau et al., 2018; Rapisarda et al., 2018; Reid et al., 2018), and experimental studies have examined the effects of asbestos on macrophages, T cells, and cytokine profiles (Ferro et al., 2014; Khaliullin et al., 2020; Kumagai-Takei et al., 2020). An emerging area of research is exploring the effects on inflammation and autoimmune disease of a severe and sudden high-intensity mixture of respirable exposures, or the combination of these exposures with highly stressful conditions, such as experienced during the September 11, 2001, World Trade Center attacks and during natural disasters such as earthquakes and tsunamis. With respect to the respirable occupational exposures noted above, the question of relevant exposure level—the combination of intensity and duration needed to induce autoimmune disease—and whether current occupational exposure limits are sufficient to prevent adverse autoimmune disease, is a crucial question that further research needs to address.
Cigarette smoking is a well-studied risk factor for many forms of cancer, cardiovascular disease, and other conditions, and for some autoimmune diseases there are relatively strong data supporting an association, including evidence of a dose–response relationship between the intensity and duration of cigarette smoking and the likelihood of developing an autoimmune disease. This is seen, for example, with rheumatoid arthritis, particularly in people with rheumatoid factor or anti-citrullinated protein peptide antibodies (Costenbader and Karlson, 2006; Liu et al., 2019), and Graves’ disease, particularly in people with Graves’ ophthalmopathy (Vestergaard, 2002). Studies have identified weaker associations between current smoking and SLE and multiple sclerosis (Belbasis et al., 2015; Parisis et al., 2019). Somewhat paradoxically, smoking is associated with an increased risk and severity of Crohn’s disease, but with a reduced risk of developing ulcerative colitis (Piovani et al., 2021); smoking cessation has been shown to increase the risk of developing ulcerative colitis in the Western world (Ananthakrishnan et al., 2020). Studies in Western cohorts,
however, have consistently demonstrated that smoking increases the risk and severity of Crohn’s disease (Ananthakrishnan et al., 2020). Research has also found that smoking is associated with severity, progression, risk of flares, or treatment effectiveness in several other autoimmune diseases (Costenbader and Karlson, 2006; Hedström et al., 2021; Parisis et al., 2019; Simmons et al., 2021).
Other Occupational and Environmental Exposures
Studies have examined the relationship of exposure to the broad category of solvents and the development of several autoimmune diseases. In 2010, an expert panel concluded that evidence from epidemiologic studies provided “confidence that solvent exposure contributes to the development of systemic sclerosis (scleroderma),” while the panel considered a link between solvent exposure and multiple sclerosis to be “likely but requiring confirmation” (Miller et al., 2012; Parks et al., 2014). Regarding specific solvents, studies of trichloroethylene exposure in animal models found disease acceleration in lupus-prone mice, markers of inflammatory response, and the development of autoimmune hepatitis, inflammatory skin lesions, and alopecia. Human studies have produced similar findings in workers exposed to trichloroethylene (Cooper et al., 2009). There is a need for further research focusing on the effects of specific solvents in experimental and epidemiologic studies.
Endocrine-disrupting chemicals—bisphenol A (BPA), a variety of phthalates, polybrominated flame retardants, perchlorate, perfluorinated chemicals, polychlorinated biphenyls, dioxins, and some types of pesticides and herbicides, among others—are another broad category of compounds of particular relevance to the general population because of the potential for exposure through environmental contamination as well as through food, food packaging, and personal care and home products (Diamanti-Kandarakis et al., 2009). Exposure to endocrine-disrupting chemicals in utero also carries the potential to affect DNA methylation and genomic imprinting; this early-life exposure can carry risk for disease even in adulthood (Robles-Matos et al., 2021). Biomonitoring data from the National Health and Nutrition Examination Survey have revealed a high prevalence and widely varying levels of exposure to many of these compounds in the U.S. population (EPA, 2021). Endocrine-disrupting chemicals such as phenols (e.g., BPA), parabens, and phthalates have been shown to influence sex hormone activity and alter immune responses in animals and may influence sex differences in autoimmune diseases as has been found for viral myocarditis in mice (Bruno et al., 2019; Castro-Correia et al., 2018; Edwards et al., 2018; Popescu et al., 2021). Compounds within this category may exhibit estrogenic, antiandrogenic, or thyroid
hormone effects, including disruption to hormone binding, synthesis, secretion, transport and metabolism. There is a growing body of research pertaining to effects of specific compounds or classes of chemical compounds on immune function (Nowak et al., 2019), but there has been relatively little focused research to date within the context of autoimmunity or autoimmune diseases.
Dietary Exposures
As Chapter 3 described, autoimmune diseases share several diet-related risk factors (Table 4-1), suggesting common etiologies and mechanisms. Several of the shared dietary risk factors, including breast milk, gluten, vitamin D, omega-3 fatty acids, and sugars, play a significant role in shaping the gut microbiome (Hamilton-Williams et al., 2021), and researchers theorize that these dietary components may interact with the microbiome to stimulate a mucosal immune response. Alternatively, or in addition, dietary intake influences the inflammatory state of the body, which may also play a role in the development of autoimmune diseases. Although vitamin D is considered a vitamin, it is actually a steroid hormone that binds to the vitamin D receptor with downstream transcriptional regulation of many genes (Tintut and Demer, 2021), and similar to other steroid hormones, vitamin D strongly influences immune responses (Park and Han, 2021).
Research has also found that proinflammatory dietary factors, such as red meat and sugar-sweetened beverages, are associated with increased risk of rheumatoid arthritis, inflammatory bowel disease, and type 1 diabetes, whereas anti-inflammatory dietary factors, such as omega-3 fatty acids, have been associated with decreased risk of type 1 diabetes and rheumatoid arthritis. The recently published Vitamin D and Omega 3 Trial found that supplementation with vitamin D led to a 22 percent reduction in all incident autoimmune diseases, and supplementation with marine omega-3 fatty acids (fish oil) with or without vitamin D led to a (nonsignificant) 15 percent reduction in all incident autoimmune diseases during 5 years of randomized follow-up of more than 25,800 older adults from the general population (Hahn et al., 2022). The study was not adequately powered to examine the effect of supplementation on individual autoimmune diseases. The list of shared dietary risk factors in Table 4-1 was limited to those observed in prospective studies. It is likely that additional prospective investigations of diet in other autoimmune diseases would uncover more shared dietary risk factors, thereby providing clues into shared etiologies and mechanisms. Additionally, systematic collection of longitudinal data on dietary exposures would enable examination of their role in development, progression, and mitigation of autoimmune diseases.
TABLE 4-1 Shared Dietary Risk Associations Across Autoimmune Diseases
| Dietary Factor | Increased risk for: | ||||||
|---|---|---|---|---|---|---|---|
| T1D | Celiac Disease | UC | Crohn’s Disease | RA | SLE | MS | |
| No breast feeding in infancy | |||||||
| Lower vitamin D intake/25OHD level | |||||||
| Lower omega-3 fatty acid intake/status | |||||||
| Higher gluten intake in early childhood | |||||||
| Higher sugar/sugar-sweetened beverage intake | |||||||
| Higher meat/animal protein intake | |||||||
NOTES: Dietary factors listed in the left column are associated with autoimmune diseases listed across the top as indicated by a filled black box.
MS, multiple sclerosis; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; T1D, type 1 diabetes; UC, ulcerative colitis; 25OHD, 25-hydroxyvitamin D.
SOURCES: No breast feeding: T1D (Lund-Blix et al., 2017), UC and Crohn’s disease (Barclay et al., 2009; Klement et al., 2004; Ng et al., 2015). Lower omega-3 fatty acids: T1D (Norris et al., 2020), RA (Di Giuseppe et al., 2014; Gan et al., 2016, 2017a,b). Lower Vitamin D/25OHD: T1D (Miettinen et al., 2020), Crohn’s disease (Ananthakrishnan et al., 2012), SLE (Hassanalilou et al., 2018; Young et al., 2017), MS (Munger et al., 2006). Higher gluten: celiac disease (Andrén Aronsson et al., 2019), T1D (Norris et al., 2020). Sugar/sugar-sweetened beverage intake: T1D (Norris et al., 2020), UC (Racine et al., 2016), RA (Hu et al., 2014). Higher meat/animal protein: Crohn’s disease and UC (Hou et al., 2011; Jantchou et al., 2010), RA (Pattison et al., 2004).
The preceding discussion of environmental factors provides a foundation for considering potential approaches to preventing the occurrence of autoimmune diseases. Some approaches are based outside of the health care system such as advertising efforts to discourage smoking or work-site interventions to reduce levels of respirable silica exposure. Other approaches rely on health care provider interactions with individuals, including efforts to identify people at high(er) risk for developing autoimmune diseases; monitor biomarkers of disease development such as type 1 diabetes-linked antibodies; apply exposure interventions; and use pharmaceutical agents that research shows can reduce disease risk, as seen in ongoing studies for type 1 diabetes and rheumatoid arthritis.
Finding: Environmental factors such as infection, occupational exposure to respirable silica, solvents, and diet have been implicated in multiple autoimmune diseases, and the same individual environmental factor may also be associated with multiple autoimmune diseases. Evidence suggests that genetic predisposition may work in concert with environmental exposures to cause autoimmune disease; moreover, there is evidence to suggest that both genetic and environmental exposure must be present in order for disease to develop. The role of environmental exposures in disease progression and flares is an important area requiring additional research.
Conclusion: Research is needed to define the type and duration of environmental exposures associated with development and progression of autoimmune diseases, including disease flares, both for specific diseases and across diseases. Similarly, further research is needed to identify the permutation of genes and environmental exposures that lead to autoimmune diseases. In addition, investigations into how dietary antigens and other environmental exposures interact with the microbiome are needed.
Inflammation, Autoantibodies, and Mechanisms of Autoimmune Disease
The presence of autoantibodies is a common feature of autoimmune diseases, as is the presence of inflammatory cells such as mononuclear phagocytes, autoreactive T lymphocytes and autoantibody-producing B cells. Autoantibodies play a key role in disease diagnosis, and they may be pathogenic in some cases when they bind to self-tissues, activate the complement cascade, and instigate cell destruction and/or clearing cell debris (Burbelo et al., 2021; Daha et al., 2011). They can also interact with and alter the function of cell-surface receptors, as is the case in myasthenia
gravis and Graves’ disease (Chain et al., 2020; George et al., 2021; Kojima et al., 2021).
Immune system cells induced during inflammation can play a major pathogenic role in many autoimmune diseases, either by killing cells directly or by releasing cytotoxic cytokines, prostaglandins, or reactive nitrogen or oxygen intermediates. At the same time, other immune system cells, such as suppressor or regulatory T cells, function as a counterbalance in controlling inflammation and autoimmune responses by killing self-reactive T cells and releasing anti-inflammatory cytokines. Disrupting the regulation of self-reactive T cells and autoantibody production by regulatory T cells may result in the development of autoimmunity. Similarly, breaking tolerance against self-antigens may trigger the development of autoimmune disease (Abbas et al., 2004; Marx et al., 2021; Nelson, 2004). Self-tolerance develops through carefully regulated mechanisms, and defects in one or more of these mechanisms may lead to autoimmune disease development.
Inoculating animals with self-antigen in combination with adjuvants that activate the immune response enables investigators to produce animal models that recreate many of the features of human autoimmune diseases (see Chapter 3 for more information on animal models for specific autoimmune diseases). Injecting adjuvants without self-antigen or self-antigen without adjuvants does not usually result in the development of autoimmune disease. Thus, in the context of presentation of self-antigen that occurs presumably as a result of viral infection or chemical exposure, it is necessary to activate the innate immune system in order to develop the adaptive immune response (i.e., a T cell and B cell response) that progresses to autoimmune disease. Adjuvant-stimulated collagen-induced rheumatoid arthritis and cardiac myosin-induced myocarditis are examples of this type of animal model (Fontes et al., 2017). The self-antigens that are responsible for causing autoimmune disease have been identified for a number of autoimmune diseases including cardiac myosin for myocarditis (Myers et al., 2013), myelin basic protein or myelin oligodendrocyte glycoprotein for multiple sclerosis (Martinsen and Kursula, 2022; Yannakakis et al., 2016), and collagen for rheumatoid arthritis (Allen et al., 2019; Liang et al., 2019). However, the autoantigens associated with most autoimmune diseases remain unknown. Identifying new autoantigens is necessary in order to develop new animal models of disease, to better detect autoantibodies and autoreactive T cells, and to develop new and more effective therapies.
Increased production of the proinflammatory cytokines TNF and IL-1b are also involved in the development of some autoimmune diseases. Injecting strains of mice that are genetically susceptible to developing autoimmunity with either of these cytokines, or with certain adjuvants
(i.e., lipopolysaccharide that binds to TLR4 releasing IL-1β) that stimulate their production, can accelerate autoimmune disease. Repeating the same experiment with genetically resistant mouse strains can break self-tolerance and lead to autoimmunity (Lane et al., 1992, 1993). This suggests that environmental factors can overcome genetic resistance to developing autoimmune disease.
Disease Predictors
A number of biomarkers obtained from serum, tissues, or other sources have been used to predict the development of an autoimmune disease, to confirm diagnosis, or to predict the outcome of disease, such as autoantibodies, cytokines, microRNAs, and T cell–B cell interactions.
Autoantibodies are common in autoimmune diseases, and, while their role in the disease pathogenesis is not always clear, detecting them in clinical specimens may aid in diagnosing or, in some cases, predicting disease. In type 1 diabetes, perhaps the most studied autoimmune disease, autoantibodies that target intracellular autoantigens prior to the patient developing autoimmune pathology are detectable many years prior to the onset of clinical symptoms (Burbelo et al., 2021; Leslie et al., 2001). The risk of developing type 1 diabetes within 5 years increases from approximately 10 percent when a blood sample shows one autoantibody to 60 to 80 percent when it shows three autoantibodies (Notkins, 2004; Ziegler et al., 2013). In SLE, autoantibodies can be detected in blood samples taken years before the clinical features of the disease appear (Arbuckle et al., 2003), and researchers have proposed that detecting autoantibodies or other measures of inflammation may be predictors of disease (Lambers et al., 2021). Similarly, studies have detected autoantibodies up to 20 years before diagnosis of Sjögren’s disease (Theander et al., 2015), and years in advance of clinical manifestations or diagnosis of other autoimmune diseases, including rheumatoid arthritis, systemic sclerosis, celiac disease, thyroid disease, and others (Burbelo et al., 2021; Leslie et al., 2001).
The ability to reliably predict the development of autoimmune diseases is an opportunity to intervene to prevent or delay development of clinical manifestations of disease and to improve outcomes. New, exciting data are now available for many autoimmune diseases, some more advanced than others, and additional studies are needed to better define the complex components driving risk to developing disease.
In type 1 diabetes, research has included focusing on genetic risk factors, exposures in infancy or before birth, and development of autoantibodies in the first years of life (Anand et al., 2021; Bonifacio et al., 2021; Frederiksen et al., 2013; Jacobsen et al., 2015; Krischer et al., 2019; Ng et
al., 2022; Ziegler et al., 2013). Such studies have made predicting disease development reliable enough to justify preventive intervention in those at high risk. The Global Platform for the Prevention of Autoimmune Diabetes Primary Oral Insulin Trial is randomizing infants with a > 10 percent risk for developing multiple autoantibodies, which in turn predicts high risk for type 1 diabetes, to receive daily oral insulin or placebo. To determine whether the intervention prevents development of autoantibodies or progression to diabetes, investigators will follow the infants up to 7 years (Ziegler et al., 2019). Because the trial participants have not yet developed evidence of autoimmunity but are at risk for doing so, it is a primary prevention trial (Primavera et al., 2020). In a secondary prevention trial, nondiabetic relatives of individuals with type 1 diabetes who were at high risk for development of disease (defined as having two or more autoantibodies and dysglycemia but not overt diabetes) were randomized to a T cell-targeting therapy or placebo and monitored for the progression to clinical diagnosis of diabetes (Herold et al., 2019). Treatment delayed progression to diagnosis of diabetes. Given that treatment is not risk free, justification for its use requires highly reliable prediction of high-risk individuals.
Similar studies, although less advanced, are in progress to define predictive measures and preemptive interventions for rheumatoid arthritis (Deane and Holers, 2021). Genetic risk scores and certain autoantibodies inform the predictive power; documented dietary and infectious exposures can enhance prediction and provide targets for intervention (Primavera et al., 2020). In addition, recent data in the study of SLE indicate that antibody to Ro52 may predict which pre-diagnosis patients will progress to diagnosable disease (Munoz-Grajales et al., 2021).
Cytokine profiles can also be used as biomarkers. For example, the type I, II, and III interferon (IFN) cytokines are involved in SLE (Idborg and Oke, 2021), and most patients with SLE have an augmented expression of IFN-regulated genes, termed an “IFN gene signature,” with type I IFNs contributing most to the signature (Capecchi et al., 2020; Ramaswamy et al., 2021). Circulating levels of IFN-α correlate with disease activity and potentially could be used as a disease biomarker (Idborg and Oke, 2021). However, levels of IFN-γ have been shown to rise years before both type I IFN activation and clinical presentation of SLE. In addition, the patterns of cytokine increases vary with different clinical manifestations of SLE. Further research in this area will likely yield novel biomarkers.
Another potential biomarker is to use a combination of cytokines (soluble ST2, IL-6), immune cell responses (Th17), and micro RNAs to diagnose and/or predict worse outcome in myocarditis. Soluble ST2 is a cytokine in the TLR4/IL-1R family that has been found to be elevated in men and male mice with myocarditis and to be associated with worse
outcome (Coronado et al., 2019). The cytokines IL-1β and IL-6, necessary for the development of Th17-type immune responses, have been found to be elevated in men with myocarditis where Th17 responses were found to be associated with poor outcome, but only in men (Myers et al., 2016). In addition, a microRNA associated with Th17 cells was identified in viral and autoimmune animal models of myocarditis that enabled researchers to distinguish myocarditis from other forms of inflammatory heart disease (Blanco-Domínguez et al., 2021). These immune pathways (TLR4/IL-1β/IL-6/Th17) are common to many autoimmune diseases and indicate the importance of identifying common pathogenic mechanisms that may reveal new biomarkers and therapeutic strategies to prevent disease.
Many autoimmune diseases involve pathologic T cell–B cell interactions, and more specifically, implicate PD-1hi CXCR5-T peripheral helper cells, which suggests that these T helper cells might be used as biomarkers of disease progression and as a target for therapy (Marks and Rao, 2022). Biomarkers also may be used to predict disease-activity changes such as flares of disease, as was recently reported using cytokines to identify renal and non-renal disease flares in individuals with SLE (Ruchakorn et al., 2019).
The most effective disease predictors will likely need to be combinations of biomarkers and/or algorithms rather than an individual biomarker.
Finding: Autoantibodies and other biomarkers are associated with autoimmune diseases, and the presence of autoantibodies has been detected before, and sometimes long before, clinical disease develops. An increase in proinflammatory cells, cytokines, and signaling pathways has been implicated in the development of autoimmune disease.
Conclusion: Continued research is needed to identify traditional and novel biomarkers, such as autoantibodies, cytokines, and genetic risk factors, of autoimmune disease in order to predict, intervene to delay or modify outcomes, and possibly even prevent development of clinical illness.
Sex Differences
Most autoimmune diseases display a sex difference in incidence and severity of disease (Cooper and Stroehla, 2003). These differences may result from the differential effects of sex hormones; microchimerism, which is the presence of two genetically distinct cell populations in the same individual; specific genes on the X or Y chromosomes; X chromosome inactivation; and sex differences in the body’s response to environmental factors. Of these, researchers consider sex hormones to be a
major contributor to the sex differences in autoimmune diseases because of their essential role in the normal physiologic function of cells, tissues, and organs and their well-defined and extensive role in regulating the immune system in normal and pathological conditions. Sex steroid hormone receptors are expressed in or on the surface of immune cells (Buskiewicz et al., 2016), and those expressed on the surface in particular influence the immune response, as they likely contribute to immune signaling by innate and adaptive immune receptors in a sex-specific manner (Benten et al., 1999a,b; Schlegel et al., 1999).
Estrogen activates B cells, resulting in increased antibody and autoantibody responses to infection, vaccines, and autoantigens (Fischinger et al., 2019). In contrast, androgens such as testosterone and dehydroepiandrosterone decrease B cell maturation, reduce B cell synthesis of antibody, and suppress autoantibody production in humans with SLE, for example (Cook, 2008; Flanagan et al., 2017; Gubbels Bupp and Jorgensen, 2018; Regitz-Zagrosek et al., 2008). Estrogen’s ability to increase autoantibodies and T and B cell responses, which are adaptive immune system activities, could play a key role in the increased incidence and severity of many autoimmune diseases that occur more often in women than men, such as autoimmune thyroiditis, Sjögren’s disease, SLE, and systemic sclerosis (Cooper and Stroehla, 2003; Fairweather et al., 2012), while androgen’s ability to increase Toll-like receptors, mast cells, and macrophages, which are innate immune system activities, may drive autoimmune diseases in men (Fairweather et al., 2008; Gubbels Bupp and Jorgensen, 2018). However, while there is evidence that estrogen influences immune signaling pathways pivotal in autoimmune disease and that it affects antibody and autoantibody levels in autoimmune disease in different animal and tissue culture models with normal and diseased cells, the fact that estrogen levels rise substantially during pregnancy and that autoimmune disease activity does not increase during pregnancy, and in some diseases such as rheumatoid arthritis, decreases during pregnancy, suggests that more research is needed to understand the role of estrogen in autoimmune disease (Moulton, 2018). Additionally, other hormones that alter the immune response besides estrogen are released during pregnancy such as progesterone (Desai and Brinton, 2019; Piccinni et al., 2021; Ziegler et al., 2018), making it more complicated to understand the role of a specific hormones in the progression of autoimmune disease.
Environmental exposures can affect sex hormone activity. For example, endocrine disruptors such as BPA have been shown to alter sex hormone expression and ratio on/in immune and other cell types in a sex-specific manner (Böckers et al., 2020; Bruno et al., 2019), and there have been few studies on the role of endocrine-disrupting chemicals in autoimmune diseases (Bruno et al., 2019).
Sex hormones are not the only factors driving sex differences in autoimmune diseases. Other factors besides estrogen that could increase autoimmune disease in women include genes expressed on X chromosomes, as appears to be the case in SLE, for example (Umiker et al., 2014), and differential responses to stress, especially early in childhood (Dube et al., 2009). The global effect of sex hormones on immune cell phenotype and function that promotes systemic or organ-specific autoimmunity is a common mechanism that occurs among different autoimmune diseases in adults. The role of onset of autoimmunity prior to puberty also requires study.
Finding: Sex chromosomes, sex hormones, and the effects of environmental exposures on sex hormones appear to play a role in differences between males and females in the occurrence and pathogenesis of autoimmune disease.
Conclusion: Additional research is needed to illuminate the role of sex chromosomes, sex hormones, and the effect of environmental exposures on sex hormones in the development of autoimmune disease as this could inform diagnosis and treatment of autoimmune diseases.
Complications, Comorbidities, and Co-occurring or Overlapping Autoimmune Diseases
People with autoimmune diseases commonly have complications, which are “sequelae or predictable consequences of a particular diagnosis” (Iezzoni, 2013); complications are considered a direct result of the autoimmune disease or its treatment and increase the risk of developing another health problem. Examples of complications that adversely affect prognosis and treatment are cardiovascular disease in patients with diabetes and colon cancer in patients with ulcerative colitis. Patients with autoimmune diseases may also suffer comorbidities, which “are diseases with unrelated etiology” (Iezzoni, 2013) that do not share causes, symptoms, or laboratory abnormalities with the autoimmune disease. A patient may also develop a co-occurring autoimmune disease, which is one that does not share symptoms or laboratory test results with the coexisting autoimmune disease. And finally, patients with autoimmune disease may suffer from an overlapping autoimmune disease, defined as the simultaneous presence of clinical and laboratory features of multiple autoimmune diseases—for example, SLE and rheumatoid arthritis, commonly known as “rhupus” (Antonini et al., 2020).
Complications of Autoimmune Diseases
Complications of autoimmune diseases (Table 4-2) can be serious. Depression and anxiety are examples of complications that occur across a majority of autoimmune disorders. The occurrence of depression and/or anxiety in the broad range of autoimmune conditions may result from the psychological effects or from the social determinants of living with an autoimmune disorder (Martz et al., 2021), but research has not ruled out the possibility that the direct central nervous system effects of rheumatoid arthritis, SLE, and multiple sclerosis may play a role in inducing depression (Bai et al., 2016; Vallerand et al., 2018, 2019). Cardiovascular disorders, stroke, lymphoma, and renal failure complicate a number of autoimmune disorders. Sometimes complications are specific to the organ targeted by the autoimmune disorder. Hepatocellular carcinoma and portal hypertension, for example, result from cirrhosis, which occurs as a consequence of primary biliary cholangitis. It is critical for clinicians to be aware of the complications of the diagnosed autoimmune disease, to undertake appropriate screening for them, and to engage patients in self-management so that patients are knowledgeable about potential complications and successful preventive strategies, and thus will bring their symptoms to medical attention (Bodenheimer et al., 2002).
Co-occurring and Overlapping Autoimmune Disease
Table 4-3 shows the likelihood of increased risk for co-occurrence of a second autoimmune illness for the autoimmune diseases examined in Chapter 3. There is one exception: risk is decreased for co-occurring multiple sclerosis and rheumatoid arthritis (Somers et al., 2009). Study of the patterns of co-occurrence may inform knowledge of pathophysiology and mechanistic pathways for all autoimmune illnesses. For instance, both genetic susceptibility and environmental agents, including diet, likely play a role in the pathogenesis of autoimmune diseases, but how they affect clinical phenotype is unknown. As with complications, it is critical for clinicians and patients to be aware of the symptoms of a second autoimmune disease when the patient has a diagnosed autoimmune disease that has a higher association with one or more other autoimmune diseases.
Finding: Co-occurring and overlapping autoimmune diseases are common; patients in these subsets differ from those with a single autoimmune disease.
TABLE 4-2 Complications and Comorbidities Exhibited by More Than One Autoimmune Disease
| Depression and/or Anxiety | Cardiovascular Disease | Stroke | Maternal/Fetal Complications | Peripheral Neuropathy and/or Other Neurologic Complications | Cancera | Renal Complications | Eye Damage or Disease | Susceptibility to Infectionb | |
|---|---|---|---|---|---|---|---|---|---|
| Sjögren’s Disease | X | X | X | X | X | X | X | X | |
| Systemic Lupus Erythematosus | X | X | X | X | X | X | X | X | X |
| Antiphospholipid Syndrome | X | X | X | X | X | X | X | X | |
| Rheumatoid Arthritis | X | X | X | X | X | X | X | ||
| Psoriasis | X | X | X | X | |||||
| Inflammatory Bowel Disease | X | X | X | X | X | X | |||
| Celiac Disease | X | X | X | X | |||||
| Primary Biliary Cholangitis | X | Xc | Xc | ||||||
| Multiple Sclerosis | X | X | X |
| Type 1 Diabetes | X | X | X | X | X | X | X | X | |
| Autoimmune Thyroid Disease | X | X | X | X | X | X |
a Susceptibility to cancer may be the result of autoimmune disease or autoimmune disease treatment.
b Susceptibility to infection may be the result of autoimmune disease or autoimmune disease treatment.
c In individuals with primary biliary cholangitis who have cirrhosis.
SOURCE: Chapter 3.
TABLE 4-3 Autoimmune Diseases That Commonly Co-occur
| Co-occurring Autoimmune Disease | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Index Autoimmune Disease | Sjögren’s Disease | SLE | APS | RA | Psoriasis | IBD | Celiac Disease | PBC | MS | T1D | AITD | |
| Sjögren’s disease | X | X | X | X | ||||||||
| SLE | X | X | X | X | ||||||||
| APS | X | |||||||||||
| RA | X | X | X | * | X | X | ||||||
| Psoriasis | X | |||||||||||
| IBD | X | X | ||||||||||
| Celiac Disease | X | X | X | X | X | |||||||
| PBC | X | |||||||||||
| MS | X | X | X | |||||||||
| T1D | X | X | ||||||||||
| AITD | X | X | X | X | ||||||||
* Inverse association between rheumatoid arthritis and multiple sclerosis (Somers et al., 2009).
NOTES: Choose an index disease in the far-left column and read horizontally across the row to identify co-occurring diseases. AITD, autoimmune thyroid diseases; APS, antiphospholipid syndrome; IBD, inflammatory bowel disease; MS, multiple sclerosis; PBC, primary biliary cholangitis; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; T1D, type 1 diabetes.
SOURCE: Chapter 3.
Conclusion: Greater focus on patients with co-occurring and overlapping autoimmune diseases may provide insight into the pathophysiology and mechanistic pathways of autoimmune disease, better characterize this type of illness, and lead to better patient care.
Common Mechanistic Pathways as Therapeutic Targets
Despite the considerable variation in clinical symptoms or targeted organs among the different autoimmune diseases, the specific mechanisms of immune-mediated damage that leads to the different clinical pictures may share common immune pathways. As discussed above, cytokines are key mediators of immune cell effector functions, either in stimulating other immune cells to propagate disease or in directly affecting target organ cells to cause damage. The identification of specific proinflammatory signaling pathways and their cytokines as pathogenic in autoimmune diseases has resulted in targeted therapeutics that aim to block their effects. Identifying TNF as an inflammatory cytokine in synovial fluid from individuals with rheumatoid arthritis and then demonstrating that blocking TNF ameliorated disease in a mouse model of inflammatory arthritis led to the study of TNF inhibitors in the treatment of rheumatoid arthritis (Feldmann and Maini, 2010). The Food and Drug Administration (FDA) has approved TNF inhibitors for treating a number of different autoimmune diseases in adults and children including rheumatoid arthritis, juvenile idiopathic arthritis, plaque psoriasis and psoriatic arthritis, ankylosing spondylitis, inflammatory bowel disease (Crohn’s disease and ulcerative colitis), and uveitis (FDA, 2021). While TNF inhibitors have had a significant effect on the management of these autoimmune diseases for many individuals, they are not effective for everyone. Studies of those individuals who fail to respond adequately to currently available medications may help to elucidate additional cytokines or other immune pathways that can be targeted therapeutically.
As noted above, IL-1 is another cytokine that plays a pathogenic role in the inflammation associated with some autoimmune diseases. Clinical studies have shown that IL-1 inhibitors are effective and often life-saving in dampening the hyperinflammatory response in autoinflammatory diseases such as systemic juvenile idiopathic arthritis, monogenic autoinflammatory diseases such as cryopyrin-associated periodic syndromes, and cytokine storm syndromes including the hyperinflammatory response that may occur in children following SARS-CoV-2 infection (Jesus and Goldbach-Mansky, 2014; Malcova et al., 2021; Mehta et al., 2020; Tanner and Wahezi, 2020).
Type I interferons, a family of molecules that includes 17 different cytokines, have relevant roles in autoimmune diseases, as well as a key
role in anti-viral immune responses (Lazear et al., 2019). This latter role, along with the association of type I interferons with autoimmune diseases, may support the idea that viral infections participate in triggering autoimmunity. However, research has also suggested that dysregulation of the type I interferon system or stimulation of type I interferon production by endogenous triggers may be involved in the etiology of autoimmunity. One extreme example of the latter is the class of autoinflammatory diseases grouped as interferonopathies, which result from monogenic inborn errors of immunity that lead to upregulation of a type I interferon response (Sogkas et al., 2021).
Research has implicated type I interferons as playing a role in several non-monogenic autoimmune diseases based on measurements of interferons directly or, more commonly, with the detection of the interferon signature—a collection of genes that are upregulated in response to interferons. SLE is the prototypical type I interferon-associated autoimmune disease, but studies have also associated type I interferons with diseases such as Sjögren’s disease, idiopathic inflammatory myopathies, and systemic sclerosis (Jiang et al., 2020). Many of the disease risk-associated gene variants shared among autoimmune diseases are also associated with type I interferon-mediated signaling pathways (Figure 4-1). Additional support for a pathogenic role for type I interferons came from the development of autoimmune manifestations following treatment with type I interferons for cancer or viral hepatitis infections. These manifestations included effects on immune-mediated thrombocytopenia, vasculitis, autoimmune thyroid disease, autoimmune hepatitis, Raynaud phenomenon, rheumatoid arthritis, myositis, psoriasis, interstitial nephritis, autoimmune colitis, and SLE, and product inserts for type I interferon drugs noted them as potential adverse effects of treatment (FDA, 2021). Other autoimmune diseases, such as multiple sclerosis, may actually improve with type I interferon treatment, suggesting that the role of type I interferons in driving autoimmunity is not absolute (FDA, 2021).
The pathogenic role of type I interferons in multiple autoimmune diseases or monogenetic autoinflammatory diseases has led to developing treatments to target the type I interferon pathway; these include blocking type I interferons or the type I interferon receptor directly, or through small molecule inhibitors such as the Janus kinase (JAK) inhibitors that target key signaling components of the type I interferon signaling pathway. FDA has approved JAK inhibitors for the treatment of multiple autoimmune diseases including rheumatoid arthritis, juvenile idiopathic arthritis, psoriatic arthritis, and ulcerative colitis (Harrington et al., 2020; O’Shea, 2021; Troncone et al., 2020), and clinical trials are ongoing in other autoimmune diseases such as systemic sclerosis, Sjögren’s disease, SLE, idiopathic inflammatory myopathies, alopecia areata, vitiligo, psoriasis,
Crohn’s disease, spondyloarthritis/ankylosing spondylitis, and vasculitis (NIH, 2021). The JAK family comprises four different kinases (JAK1, JAK2, JAK3, tyrosine kinase 2) that may each be blocked somewhat selectively by different drugs. Different JAKs or combinations of JAKs are used by different cytokine receptors to mediate signaling, so JAK inhibitors may be used to block signaling through multiple different inflammatory cytokine receptors as appropriate for specific diseases. Additional drugs specifically targeting other inflammatory cytokines, including but not limited to IFN-g, IL-17, IL-23, and IL-12/23 shared subunit, are in use or in development for treating multiple different autoimmune diseases that share these specific inflammatory cytokines in the disease process (Baker and Isaacs, 2018).
Further studies to identify common immune pathways in different autoimmune diseases or subsets of different diseases will continue to provide insight into potentially targetable molecules that could lead to treatments for multiple different autoimmune diseases.
Finding: Specific mechanisms of immune-mediated damage that lead to the different clinical phenotypes share common immune pathways. Identifying specific proinflammatory signaling pathways and their cytokines as pathogenic in autoimmune diseases has yielded targeted therapeutics that aim to block their effects, as with, for example, TNF inhibitors and IL-1 inhibitors.
Conclusion: Continued research is needed to identify additional immune pathways that promote or inhibit autoimmune disease. Such pathways may be common to multiple autoimmune diseases and inform drug development.
Animal Models
Animal models have played a critical role in defining common genetic and environmentally-induced mechanisms that drive the pathogenesis of autoimmune diseases as described in this chapter. Chapter 3 describes some of these animal models for the autoimmune diseases that are the focus of this report, but a detailed description of the many animal models investigators are using and the key findings and discoveries that studies using these models have produced is outside the scope of this report.
Existing animal models form a critical component of autoimmune disease research by providing mechanistic understanding of autoimmune diseases and preclinical data needed for translational research. For example, environmental causes of several human autoimmune diseases including celiac disease (Wu et al., 2021), rheumatic fever (de Loizaga and Beaton, 2021), and myocarditis (Boehmer et al., 2021) are known, and these
diseases could be used as models to accelerate identification of triggers and primary prevention for others. The ability to directly study effects of genetic modification, environmental effects, or potentially therapeutic immunomodulatory agents on the function of the immune system or the development of autoimmunity has been invaluable. Moreover, the ability to study immune cells and non-immune cells harvested directly from the target organs may provide a basis for subsequent studies in human tissues, which are often much less readily available or accessible. The committee wants to emphasize the importance of existing animal models that continue to provide valuable understanding of disease pathogenesis as well as the need to develop animal models for many autoimmune diseases where animal models do not currently exist and the need to continue to develop new animal models that answer specific and more translational research questions.
Future advances in tissue culture, such as 3D organoids, or organs-on-a-chip, which can model organs and systems such as the immune system (Park et al., 2020; Polini et al., 2019), as well as novel culture, animal, and other models, such as in silico confirmatory trials that use computer simulation to model virtual cohorts of patients (Pappalardo et al., 2019), may provide additional insight into autoimmune disease mechanisms or effective therapies.
Finding: Existing animal models are invaluable for studying mechanisms of autoimmune disease pathogenesis and providing preclinical data for translational research.
Conclusion: Developing animal models for autoimmune diseases that currently have no models or are rare diseases and continuing to develop animal models that can better answer mechanistic questions posed by translational research would be of value. Technological innovation for the creation of new models for autoimmune disease research is needed.
MAXIMIZING ADVANCES IN TECHNOLOGY
Over the past decade, the availability of innovative technologies has completely changed the research environment and has come to dominate research on autoimmune diseases. These technological advances and the knowledge they have generated include the ability to perform more complex analyses of biological systems; knowledge about genomics and epigenomics; cytokine profiling; microbiomics, proteomics, metabolomics, and the study of changes in these “omics”; and generation of artificial intelligence. Fine-mapping genetic and epigenetic causal autoimmune disease variants, for example, enables researchers to identify clustering
among autoimmune diseases (Farh et al., 2015). As an example, a genetic region containing the gene for the enzyme tyrosine kinase 2 was identified in genome-wide association studies as linked to many autoimmune diseases, including psoriasis, SLE, and Crohn’s disease (Makin, 2021). A disease-associated gene variant modifies the enzyme, which plays a role in cytokine function. Therapy to block enzyme function greatly reduced skin lesions in psoriasis, and a phase III trial in psoriasis and early-stage trials in Crohn’s disease, SLE, and psoriatic arthritis are under way.
The ability to study gene expression profiles in immune cells at the single-cell level provides insights into the complex processes within heterogeneous tissue samples (Yofe et al., 2020). Moreover, combining the transcriptomics with proteomics on a single cell level (such as with flow or mass cytometry) can provide tremendous detail of complex subsets of immune and non-immune cells contributing to the inflammatory state within affected tissue. Analyses of synovial fluid from individuals with rheumatoid arthritis compared with non-autoimmune arthritis (osteoarthritis) combined single-cell RNA sequencing with mass cytometry and identified 18 unique cell populations—including different subsets of T cells, B cells, monocytes, and fibroblasts—and discovered the specific subsets expressing genes for key inflammatory cytokines IL-6 (expressed by fibroblasts) and IL-1b (expressed by pro-inflammatory monocytes) (Zhang et al., 2019). Another study utilizing mass cytometry coupled analyses from labial minor salivary gland biopsy samples with blood samples from individuals with Sjögren’s disease to define immunopheno-types of cells from the affected organs and the peripheral blood (Mingueneau et al., 2016). The study identified different disease clusters based on profiles of cells within the affected tissue and peripheral blood including associations with disease activity parameters and potential pathogenic cells to be targeted therapeutically. In another study, single cell sequencing to define T-cell receptor gene expression to identify clones of T cells expanded within affected tissue samples of individuals with Sjögren’s disease identified common T-cell receptor sequences within and between individual samples, strongly suggesting common antigen-driven immune response between different individuals (Joachims et al., 2016). The techno-logic advances afforded by single cell RNA sequencing provides a level of detail not possible through other measures to identify specific clonotypes of T cells in the absence of known antigens even from small samples such as cells isolated from biopsy specimens routinely obtained for diagnostic purposes. The analyses of such complex data sets are continually evolving, with recent study demonstrating the utility of latent factor modeling of single cell RNA sequencing data to define new pathways active in synovial tissues of individuals with rheumatoid arthritis and in the kidneys of individuals with SLE (Palla and Ferrero, 2020). Additional single
cell RNA sequencing studies have been performed in rheumatoid arthritis, SLE, systemic sclerosis, psoriatic arthritis, spondyloarthritis, Sjögren’s disease, Kawasaki disease, multiple sclerosis, and type 1 diabetes (Kuret et al., 2021; Zhao et al., 2021). A possible avenue is to use high-throughput screening of thousands of autoantigens in parallel rather than single autoantigens or panels of single autoantigens. The continued development of new technologies and complex data-analysis methodologies will continue to gain unprecedented detailed knowledge of the immunopathologic processes in autoimmune diseases.
Gene-editing techniques are available to study the effects of insertion/deletion/alteration of genes of interest on immune cell function and disease models (Pavlovic et al., 2020). On a public health level, the use of these omics are being incorporated into defining the exposome, which will describe the effects of lifetime environmental exposures on health and disease (DeBord et al., 2016; Manrai et al., 2017).
Given the complexities of autoimmunity on multiple levels, researchers are using multi-omics approaches to provide a more comprehensive understanding of the immune response and the interplay between different components (cells, cytokines, genes, metabolism) (Chu et al., 2021). Such multi-omics approaches have demonstrated that distinct molecular clusters characterize individual systemic autoimmune diseases despite common clinical manifestations and that such clusters are similar across different systemic autoimmune diseases despite differences in clinical presentation (Barturen et al., 2021). These advanced technologies are generating datasets of increasing size and complexity, requiring more complex analyses that include systems modeling and machine learning, which have also advanced during this period and are providing additional insight into the complexities of autoimmune diseases (Brown et al., 2018).
Artificial intelligence (AI) and machine learning, for example, can be used to search electronic health records. An examination of electronic health records of 167,109 patients with rheumatoid arthritis found that the risk for lower-tract gastrointestinal perforation associated with treatment with tocilizumab and tofacitinib to be more than twice that of anti-TNF (Xie et al., 2016). These tools can be used for precision medicine, a term that encompasses determining disease treatment for individuals; prospective public health approaches that target disease prevention; and precision public health approaches to classify patients into at-risk or disease subsets for interventions (Conrad et al., 2020). In addition, phenome-wide association studies match genetic variants to corresponding phenotypes in electronic health records to gain insight into outcomes and disease risk.
Regardless of the kind of data examined or the investigative purpose, the value of output depends on the rigor of the analytic methods used
to collect and interpret it. Big data analytics involves “integration of heterogeneous data, data quality control, analysis, modeling, interpretation and validation” (Ristevski and Chen, 2018), and the skill sets needed to fulfill these needs are essential to robust research. In addition, sound use of big data includes planning for optimum usability; in the case of electronic health records, for example, this would mean to share, clean, and archive data.
A multidisciplinary approach to longitudinal data collection in diverse population-based cohorts is needed in order to provide a strong foundation for integrated basic, translational, and clinical research necessary to address the complex and challenging questions relating to development, progression, and treatment of autoimmune diseases. Immunology, genetics, and clinical expertise in a variety of specialties including environmental health, epidemiology, and data science are among the disciplines needed to optimize the design, conduct, and analysis of this research. Existing and newly developed large data sets should apply rapid improvements in big data-analysis tools, including AI and machine learning, to advance knowledge in autoimmune diseases. For example, technical advances should be harnessed to increase information on sub-populations. Technologic tools could be used, among other things, to:
- examine genetic variation via genome-wide association studies;
- examine health disparities including the molecular underpinnings of differences in disease across race and ethnicity and how these might impact treatment;
- study environmental triggers for disease onset, disease flares, disease progression, and poor outcomes;
- elucidate immunophenotype immune cell pathways;
- identify metabolic pathways;
- decipher within and across disease-homogenous subsets; and
- compare persons with minimal disease or disease that has not yet manifested with those with severe disease or poor outcomes in order to expose key pathways to target in autoimmunity.
New and evolving technologies are supporting and advancing basic science, and translational research, to which the National Center for Advancing Translational Sciences is dedicated, is developing and using innovative technologies and methodologies to better search and interpret basic science findings in order to identify opportunities for enhancing diagnostics and therapeutics. The accruing knowledge will guide clinical research, from informing the types of samples collected and tests performed in observational studies to the design of intervention trials.
Finding: Advances in technology are rapidly providing novel insights into disease pathogenesis including autoimmune diseases. Multi-omics approaches are providing unique information about genetic and immune profiles that contribute to individual autoimmune diseases as well as revealing common pathogenic mechanisms between autoimmune diseases.
Conclusion: Continued development of innovative technologies will be important in providing breakthroughs in understanding mechanisms of autoimmune disease and may reveal new biomarkers that predict, diagnose, or prevent disease.
SUMMARY AND RESEARCH IMPLICATIONS
Characteristics that distinguish autoimmune diseases from each other and those that are common across autoimmune diseases provide valuable information about the pathogenesis of disease. These similarities and differences provide insights to better understand the epidemiology of disease, early predictors/biomarkers of disease, diagnosis, whether a disease is predicted to primarily affect an organ or have a systemic distribution, the occurrence of a flare, and the type of immune response that is important in driving disease, for example. The descriptions of common features across autoimmune diseases that this chapter provides lead to several research opportunities.
One implication of considering autoimmune diseases according to commonalities is that considering autoimmune diseases as a group according to common mechanisms of disease might provide novel insights beyond those gained by the more common approach that focuses on the affected organ or system. Examples of mechanistic themes that are common across autoimmune diseases that this chapter discusses include sex differences in incidence, severity, and type of immune response and the role of type I interferons in increasing autoimmune pathology (Barrat et al., 2019; Psarras et al., 2017). The current tendency of many researchers to focus on the organ or system may stem from the National Institutes of Health’s (NIH’s) organization of study sections based on that scheme. Animal models that better represent the clinical pathology and course of disease would enable studies that explore these common mechanisms. As a personalized medicine approach to health care (i.e., genomics/pharmacogenomics) becomes more common in medical practice, it becomes increasingly important to understand common mechanisms of autoimmune disease given that autoimmune disease may influence other coexisting inflammatory illnesses that are increasingly being considered as autoinflammatory diseases, including cardiovascular diseases such as
atherosclerosis. In addition, it is common for a patient with one autoimmune disease to have another autoimmune disease.
Many of the genes that confer susceptibility to autoimmune disease and that different autoimmune diseases share regulate the immune response, and some autoimmune diseases with very different clinical characteristics share genetic variants (Sogkas et al., 2021) (see Figure 4-1 and Figure 4-2). Fine-mapping genetic and epigenetic causal autoimmune disease variants can detect clustering among autoimmune diseases (Farh et al., 2015). Studying the function of genetic variant-related immune pathways will be critical to revealing the genotype–phenotype relationships that could provide targets for therapies. Polygenic risk scores are useful, but still have limitations, one being that the genomics data they are based on would likely need to include more diverse populations in order to be more broadly relevant.
The number and type of autoantibodies predict progression to autoimmune disease, but research is lacking in this area across autoimmune diseases. Insight can be gained by better characterizing autoantibodies that exist in many autoimmune diseases as well as those that are distinct for a particular disease and using those similarities and differences to better diagnose disease, predict the risk of developing an autoimmune disease, and predict the progression of disease. Cytokine profiles and microRNAs, for example, have demonstrated similar opportunities (Blanco-Domínguez et al., 2021; Idborg and Oke, 2021), and research on these and other novel biomarkers is called for.
For some autoimmune diseases, Black, Hispanic, and Indigenous people have an increased risk and greater severity of disease (Izmirly et al., 2021), while for other diseases, the highest risk occurs among White populations (Dabelea et al., 2014). Research pertaining to racial and ethnic patterns needs to fully address the spectrum of experiences represented by these findings, including living and work environments, social and economic factors, and a wide variety of stressors. To date, few studies focusing on race and ethnicity have used this lens.
Although sex differences in autoimmune diseases have been known and studied for some time, large gaps in defining sex and age differences and similarities remain. The field would benefit from research on sex differences in epidemiology, risk-factor identification and assessment, diagnostic criteria, coexisting illnesses, outcomes, and other topics. The committee emphasizes that sex differences need to be examined within the context of age because of the significant changes in sex hormone levels that occur across the lifespan in females and males. There is a gap in research across the lifespan for most autoimmune diseases.
An understanding of potential environmental causes of autoimmune diseases that is based on a systematic study of exposures including
dietary factors over time is needed to devise effective approaches to prevent and/or treat disease, including disease flares. Gene and environment interactions are also understudied. Twin concordance studies indicate the importance of both factors in contributing to autoimmune disease (Brix et al., 2001; Nistico et al., 2012; Ulff-Møller et al., 2018; Willer et al., 2003) research to better understand this relationship is needed. Current research indicates that environmental factors drive disease in individuals with genetic susceptibility (Bjornevik et al., 2022; Mehri et al., 2020; Robles-Matos et al., 2021; Vestergaard, 2002), yet the mechanisms behind the relationship remain unclear. It will be important to establish study sections that support research that examines interactions between genes and multiple environmental factors such as infections and chemicals. Currently an unintentional and artificial barrier exists, with grants for environmental topics going to the National Institute of Environmental Health Sciences, for example, and grants for infectious diseases going to the National Institute of Allergy and Infectious Diseases. This type of research may be important for efforts to develop effective intervention strategies to prevent development of disease in people who are at higher risk for developing disease.
Based on the known sex differences in disease for most autoimmune diseases and the well-described effect of sex hormones on immune cells and antibodies (Cook, 2008; Fischinger et al., 2019; Flanagan et al., 2017; Gubbels Bupp and Jorgensen, 2018; Regitz-Zagrosek et al., 2008), an area that lacks clinical and basic research studies includes the effect of exogenous hormone endocrine disruptors such as BPA and bisphenol S, which replaced BPA in plastic drinking bottles, on autoimmune disease. Also needed are studies on other hormone influencing factors such as those in diet—soy, for example—and medications such as birth control pills.
That research has linked several different infectious agents and chemicals to one autoimmune disease and one infectious agent to several different diseases suggests that common innate immune mechanisms are critical in driving disease. More research is needed to better characterize these common pathways across multiple autoimmune diseases clinically and through basic research. A better understanding of the mechanisms that occur commonly in many autoimmune diseases could fundamentally improve understanding of the pathogenesis of autoimmune disease and benefit patients with these conditions. Current technologies and methodologies such as multi-omics approaches, have already contributed greatly to this end, and continued development of innovative tools can support and drive further advances.
REFERENCES
Abakkouy, Y., and I. Cleynen. 2021. The promise of polygenic risk scores as a research tool to analyse the genetics underlying IBD phenotypes. Journal of Crohn’s & Colitis 15(6):877–878. https://doi.org/10.1093/ecco-jcc/jjab021.
Abbas, A. K., J. Lohr, B. Knoechel, and V. Nagabhushanam. 2004. T cell tolerance and autoimmunity. Autoimmunity Reviews 3(7–8):471–475.. https://doi.org/10.1016/j.autrev.2004.07.004.
Acosta-Herrera, M., M. Kerick, D. González-Serna, C. Myositis Genetics, C. Scleroderma Genetics, C. Wijmenga, A. Franke, P. K. Gregersen, L. Padyukov, J. Worthington, T. J. Vyse, M. E. Alarcón-Riquelme, M. D. Mayes, and J. Martin. 2019. Genome-wide meta-analysis reveals shared new loci in systemic seropositive rheumatic diseases. Annals of the Rheumatic Diseases 78(3):311–319. https://doi.org/10.1136/annrheumdis-2018-214127.
Allen, R., S. Chizari, J. A. Ma, S. Raychaudhuri, and J. S. Lewis. 2019. Combinatorial, microparticle-based delivery of immune modulators reprograms the dendritic cell phenotype and promotes remission of collagen-induced arthritis in mice. ACS Applied Bio Materials 2(6):2388–2404. https://doi.org/10.1021/acsabm.9b00092.
Anand, V., Y. Li, B. Liu, M. Ghalwash, E. Koski, K. Ng, J. L. Dunne, J. Jönsson, C. Winkler, M. Knip, J. Toppari, J. Ilonen, M. B. Killian, B. I. Frohnert, M. Lundgren, A. G. Ziegler, W. Hagopian, R. Veijola, and M. Rewers, for the T1D1 Study Group. 2021. Islet autoimmunity and HLA markers of presymptomatic and clinical type 1 diabetes: Joint analyses of prospective cohort studies in Finland, Germany, Sweden, and the U.S. Diabetes Care 44(10):2269–2276. https://doi.org/10.2337/dc20-1836.
Ananthakrishnan, A. N., G. G. Kaplan, and S. C. Ng. 2020. Changing global epidemiology of inflammatory bowel diseases: Sustaining health care delivery into the 21st century. Clinical Gastroenterology and Hepatology 18(6):1252–1260. https://doi.org/10.1016/j.cgh.2020.01.028.
Ananthakrishnan, A. N., H. Khalili, L. M. Higuchi, Y. Bao, J. R. Korzenik, E. L. Giovannucci, J. M. Richter, C. S. Fuchs, and A. T. Chan. 2012. Higher predicted vitamin d status is associated with reduced risk of Crohn’s disease. Gastroenterology 142(3):482–489. https://doi.org/10.1053/j.gastro.2011.11.040.
Anderson, C. A., G. Boucher, C. W. Lees, A. Franke, M. D’Amato, K. D. Taylor, J. C. Lee, P. Goyette, M. Imielinski, A. Latiano, C. Lagace, R. Scott, L. Amininejad, S. Bumpstead, L. Baidoo, R. N. Baldassano, M. Barclay, T. M. Bayless, S. Brand, C. Buning, J.-F. Colombel, L. A. Denson, M. De Vos, M. Dubinsky, C. Edwards, D. Ellinghaus, R. S. N. Fehrmann, J. A. B. Floyd, T. Florin, D. Franchimont, L. Franke, M. Georges, J. Glas, N. L. Glazer, S. L. Guthery, T. Haritunians, N. K. Hayward, J. P. Hugot, G. Jobin, D. Laukens, I. Lawrance, M. Lemann, A. Levine, C. Libioulle, E. Louis, D. P. McGovern, M. Milla, G. W. Montgomery, K. I. Morley, C. Mowat, A. Ng, W. Newman, R. A. Ophoff, L. Papi, O. Palmieri, L. Peyrin-Biroulet, J. Panes, A. Phillips, N. J. Prescott, D. D. Proctor, R. Roberts, R. Russell, P. Rutgeerts, J. Sanderson, M. Sans, P. Schumm, F. Seibold, Y. Sharma, L. A. Simms, M. Seielstad, A. H. Steinhart, S. R. Targan, L. H. van den Berg, M. Vatn, H. Verspaget, T. Walters, C. Wijmenga, D. C. Wilson, H. J. Westra, R. J. Xavier, Z. Z. Zhao, C. Y. Ponsioen, V. Andersen, L. Torkvist, M. Gazouli, N. P. Anagnou, T. H. Karlsen, L. Kupcinskas, J. Sventoraityte, J. C. Mansfield, S. Kugathasan, M. S. Silverberg, J. Halfvarson, J. I. Rotter, C. G. Mathew, A. M. Griffiths, R. Gearry, T. Ahmad, S. R. Brant, M. Chamaillard, J. Satsangi, J. H. Cho, S. Schreiber, M. J. Daly, J. C. Barrett, M. Parkes, V. Annese, H. Hakonarson, G. Radford-Smith, R. H. Duerr, S. Vermeire, R. K. Weersma, and J. D. Rioux. 2011. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nature Genetics 43(3):246–252. https://doi.org/10.1038/ng.764.
Andlauer, T. F. M., D. Buck, G. Antony, A. Bayas, L. Bechmann, A. Berthele, A. Chan, C. Gasperi, R. Gold, C. Graetz, J. Haas, M. Hecker, C. Infante-Duarte, M. Knop, T. Kümpfel, V. Limmroth, R. A. Linker, V. Loleit, F. Luessi, S. G. Meuth, M. Mühlau, S. Nischwitz, F. Paul, M. Pütz, T. Ruck, A. Salmen, M. Stangel, J.-P. Stellmann, K. H. Stürner, B. Tackenberg, F. Then Bergh, H. Tumani, C. Warnke, F. Weber, H. Wiendl, B. Wildemann, U. K. Zettl, U. Ziemann, F. Zipp, J. Arloth, P. Weber, M. Radivojkov-Blagojevic, M. O. Scheinhardt, T. Dankowski, T. Bettecken, P. Lichtner, D. Czamara, T. Carrillo-Roa, E. B. Binder, K. Berger, L. Bertram, A. Franke, C. Gieger, S. Herms, G. Homuth, M. Ising, K. H. Jockel, T. Kacprowski, S. Kloiber, M. Laudes, W. Lieb, C. M. Lill, S. Lucae, T. Meitinger, S. Moebus, M. Muller-Nurasyid, M. M. Nothen, A. Petersmann, R. Rawal, U. Schminke, K. Strauch, H. Volzke, M. Waldenberger, J. Wellmann, E. Porcu, A. Mulas, M. Pitzalis, C. Sidore, I. Zara, F. Cucca, M. Zoledziewska, A. Ziegler, B. Hemmer, and B. Muller-Myhsok. 2016. Novel multiple sclerosis susceptibility loci implicated in epigenetic regulation. Science Advances 2(6):e1501678. https://doi.org/10.1126/sciadv.1501678.
Andrén Aronsson, C., H.-S. Lee, E. M. Hård af Segerstad, U. Uusitalo, J. Yang, S. Koletzko, E. Liu, K. Kurppa, P. J. Bingley, J. Toppari, A. G. Ziegler, J.-X. She, W. A. Hagopian, M. Rewers, B. Akolkar, J. P. Krischer, S. M. Virtanen, J. M. Norris, and D. Agardh, for the TEDDY Study Group. 2019. Association of gluten intake during the first 5 years of life with incidence of celiac disease autoimmunity and celiac disease among children at increased risk. JAMA 322(6):514–523. https://doi.org/10.1001/jama.2019.10329.
Antonini, L., B. Le Mauff, C. Marcelli, A. Aouba, and H. de Boysson. 2020. Rhupus: A systematic literature review. Autoimmunity Reviews 19(9):102612. https://doi.org/10.1016/j.autrev.2020.102612.
Arbuckle, M. R., M. T. McClain, M. V. Rubertone, R. H. Scofield, G. J. Dennis, J. A. James, and J. B. Harley. 2003. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. The New England Journal of Medicine 349(16):1526–1533. https://doi.org/10.1056/NEJMoa021933.
Bacon, L. D., J. H. Kite, Jr., and N. R. Rose. 1974. Relation between the major histocompatibility (B) locus and autoimmune thyroiditis in obese chickens. Science 186(4160):274–275. https://doi.org/10.1126/science.186.4160.274.
Bai, R., S. Liu, Y. Zhao, Y. Cheng, S. Li, A. Lai, Z. Xie, X. Xu, Z. Lu, and J. Xu. 2016. Depressive and anxiety disorders in systemic lupus erythematosus patients without major neuropsychiatric manifestations. Journal of Immunology Research 2016:2829018. https://doi.org/10.1155/2016/2829018.
Baker, K. F., and J. D. Isaacs. 2018. Novel therapies for immune-mediated inflammatory diseases: What can we learn from their use in rheumatoid arthritis, spondyloarthritis, systemic lupus erythematosus, psoriasis, Crohn’s disease and ulcerative colitis? Annals of the Rheumatic Diseases 77(2):175–187. https://doi.org/10.1136/annrheumdis-2017-211555.
Barclay, A. R., R. K. Russell, M. L. Wilson, W. H. Gilmour, J. Satsangi, and D. C. Wilson. 2009. Systematic review: The role of breastfeeding in the development of pediatric inflammatory bowel disease. Journal of Pediatrics 155(3):421–426. https://doi.org/10.1016/j.jpeds.2009.03.017.
Barrat, F. J., M. K. Crow, and L. B. Ivashkiv. 2019. Interferon target-gene expression and epigenomic signatures in health and disease. Nature Immunology 20(12):1574–1583. https://doi.org/10.1038/s41590-019-0466-2.
Barrett, J. C., S. Hansoul, D. L. Nicolae, J. H. Cho, R. H. Duerr, J. D. Rioux, S. R. Brant, M. S. Silverberg, K. D. Taylor, M. M. Barmada, A. Bitton, T. Dassopoulos, L. W. Datta, T. Green, A. M. Griffiths, E. O. Kistner, M. T. Murtha, M. D. Regueiro, J. I. Rotter, L. P. Schumm, A. H. Steinhart, S. R. Targan, R. J. Xavier, the NIDDK IBD Genetics Consortium, C. Libioulle, C. Sandor, M. Lathrop, J. Belaiche, O. Dewit, I. Gut, S. Heath, D. Laukens, M. Mni, P. Rutgeerts, A. Van Gossum, D. Zelenika, D. Franchimont, J. P. Hugot, M. de Vos, S. Vermeire, E. Louis, the Belgian-French IBD Consortium, the Wellcome Trust Case Control Consortium, L. R. Cardon, C. A. Anderson, H. Drummond, E. Nimmo, T. Ahmad, N. J. Prescott, C. M. Onnie, S. A. Fisher, J. Marchini, J. Ghori, S. Bumpstead, R. Gwilliam, M. Tremelling, P. Deloukas, J. Mansfield, D. Jewell, J. Satsangi, C. G. Mathew, M. Parkes, M. Georges, and M. J. Daly. 2008. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nature Genetics 40(8):955–962. https://doi.org/10.1038/ng.175.
Barrett, J. C., D. G. Clayton, P. Concannon, B. Akolkar, J. D. Cooper, H. A. Erlich, C. Julier, G. Morahan, J. Nerup, C. Nierras, V. Plagnol, F. Pociot, H. Schuilenburg, D. J. Smyth, H. Stevens, J. A. Todd, N. M. Walker, S. S. Rich, and The Type 1 Diabetes Genetics Consortium. 2009. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nature Genetics 41(6):703–707. https://doi.org/10.1038/ng.381.
Barturen, G., S. Babaei, F. Català-Moll, M. Martínez-Bueno, Z. Makowska, J. Martorell-Marugán, P. Carmona-Sáez, D. Toro-Domínguez, E. Carnero-Montoro, M. Teruel, M. Kerick, M. Acosta-Herrera, L. L. Lann, C. Jamin, J. Rodríguez-Ubreva, A. García-Gómez, J. Kageyama, A. Buttgereit, S. Hayat, J. Mueller, R. Lesche, M. Hernandez-Fuentes, M. Juarez, T. Rowley, I. White, C. Marañón, T. G. Anjos, N. Varela, R. Aguilar-Quesada, F. J. Garrancho, A. López-Berrio, M. R. Maresca, H. Navarro-Linares, I. Almeida, N. Azevedo, M. Brandão, A. Campar, R. Faria, F. Farinha, A. Marinho, E. Neves, A. Tavares, C. Vasconcelos, E. Trombetta, G. Montanelli, B. Vigone, D. Alvarez-Errico, T. Li, D. Thiagaran, R. B. Alonso, A. C. Martínez, F. Genre, R. L. Mejías, M. A. Gonzalez-Gay, S. Remuzgo, B. U. Garcia, R. Cervera, G. Espinosa, I. Rodríguez-Pintó, E. D. Langhe, J. Cremer, R. Lories, D. Belz, N. Hunzelmann, N. Baerlecken, K. Kniesch, T. Witte, M. Lehner, G. Stummvoll, M. Zauner, M. A. Aguirre-Zamorano, N. Barbarroja, M. C. Castro-Villegas, E. Collantes-Estevez, E. d. Ramon, I. D. Quintero, A. Escudero-Contreras, M. C. F. Roldán, Y. J. Gómez, I. J. Moleón, R. Lopez-Pedrera, R. Ortega-Castro, N. Ortego, E. Raya, C. Artusi, M. Gerosa, P. L. Meroni, T. Schioppo, A. D. Groof, J. Ducreux, B. Lauwerys, A.-L. Maudoux, D. Cornec, V. Devauchelle-Pensec, S. Jousse-Joulin, P.-E. Jouve, B. Rouvière, A. Saraux, Q. Simon, M. Alvarez, C. Chizzolini, A. Dufour, D. Wynar, A. Balog, M. Bocskai, M. Deák, S. Dulic, G. Kádár, L. Kovács, Q. Cheng, V. Gerl, F. Hiepe, L. Khodadadi, S. Thiel, E. d. Rinaldis, S. Rao, R. J. Benschop, C. Chamberlain, E. R. Dow, Y. Ioannou, L. Laigle, J. Marovac, J. Wojcik, Y. Renaudineau, M. O. Borghi, J. Frostegård, J. Martín, L. Beretta, E. Ballestar, F. McDonald, J.-O. Pers, and M. E. Alarcón-Riquelme. 2021. Integrative analysis reveals a molecular stratification of systemic autoimmune diseases. Arthritis & Rheumatology 73(6):1073–1085. https://doi.org/10.1002/art.41610.
Bates, M. A., C. Brandenberger, I. Langohr, K. Kumagai, J. R. Harkema, A. Holian, and J. J. Pestka. 2015. Silica triggers inflammation and ectopic lymphoid neogenesis in the lungs in parallel with accelerated onset of systemic autoimmunity and glomerulonephritis in the lupus-prone NZBWF1 mouse. PLOS ONE 10(5):e0125481.
Belbasis, L., V. Bellou, E. Evangelou, J. P. A. Ioannidis, and I. Tzoulaki. 2015. Environmental risk factors and multiple sclerosis: An umbrella review of systematic reviews and meta-analyses. The Lancet Neurology 14(3):263–273. https://doi.org/10.1016/s1474-4422(14)70267-4.
Benten, W. P., M. Lieberherr, G. Giese, C. Wrehlke, O. Stamm, C. E. Sekeris, H. Mossmann, and F. Wunderlich. 1999a. Functional testosterone receptors in plasma membranes of T cells. FASEB Journal 13(1):123–133. https://doi.org/10.1096/fasebj.13.1.123.
Benten, W. P., M. Lieberherr, O. Stamm, C. Wrehlke, Z. Guo, and F. Wunderlich. 1999b. Testosterone signaling through internalizable surface receptors in androgen receptor-free macrophages. Molecular Biology of the Cell 10(10):3113–3123. https://doi.org/10.1091/mbc.10.10.3113.
Bentham, J., D. L. Morris, D. S. C. Graham, C. L. Pinder, P. Tombleson, T. W. Behrens, J. Martín, B. P. Fairfax, J. C. Knight, L. Chen, J. Replogle, A.-C. Syvänen, L. Rönnblom, R. R. Graham, J. E. Wither, J. D. Rioux, M. E. Alarcón-Riquelme, and T. J. Vyse. 2015. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nature Genetics 47(12):1457–1464. https://doi.org/10.1038/ng.3434.
Bjornevik, K., M. Cortese, B. C. Healy, J. Kuhle, M. J. Mina, Y. Leng, S. J. Elledge, D. W. Niebuhr, A. I. Scher, K. L. Munger, and A. Ascherio. 2022. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science 375(6578):296–301. https://doi.org/10.1126/science.abj8222.
Blanco-Domínguez, R., R. Sánchez-Díaz, H. de la Fuente, L. J. Jiménez-Borreguero, A. Matesanz-Marín, M. Relaño, R. Jiménez-Alejandre, B. Linillos-Pradillo, K. Tsilingiri, M. L. Martín-Mariscal, L. Alonso-Herranz, G. Moreno, R. Martín-Asenjo, M. M. GarcíaGuimaraes, K. A. Bruno, E. Dauden, I. González-Álvaro, L. M. Villar-Guimerans, A. Martínez-León, A. M. Salvador-Garicano, S. A. Michelhaugh, N. E. Ibrahim, J. L. Januzzi, J. Kottwitz, S. Iliceto, M. Plebani, C. Basso, A. Baritussio, M. Seguso, R. Marcolongo, M. Ricote, D. Fairweather, H. Bueno, L. Fernández-Friera, F. Alfonso, A. L. P. Caforio, D. A. Pascual-Figal, B. Heidecker, T. F. Lüscher, S. Das, V. Fuster, B. Ibáñez, F. Sánchez-Madrid, and P. Martín. 2021. A novel circulating microrna for the detection of acute myocarditis. The New England Journal of Medicine 384(21):2014–2027. https://doi.org/10.1056/NEJMoa2003608.
Böckers, M., N. W. Paul, and T. Efferth. 2020. Bisphenolic compounds alter gene expression in MCF-7 cells through interaction with estrogen receptor α. Toxicology and Applied Pharmacology 399:115030. https://doi.org/10.1016/j.taap.2020.115030.
Bodenheimer, T., K. Lorig, H. Holman, and K. Grumbach. 2002. Patient self-management of chronic disease in primary care. JAMA 288(19):2469–2475. https://doi.org/10.1001/jama.288.19.2469.
Boehmer, T. K., L. Kompaniyets, A. M. Lavery, J. Hsu, J. Y. Ko, H. Yusuf, S. D. Romano, A. V. Gundlapalli, M. E. Oster, and A. M. Harris. 2021. Association between COVID-19 and myocarditis using hospital-based administrative data—United States, March 2020–January 2021. MMWR: Morbidity and Mortality Weekly Report 70(35):1228–1232. http://dx.doi.org/10.15585/mmwr.mm7035e5.
Bonifacio, E., A. Weiß, C. Winkler, M. Hippich, M. J. Rewers, J. Toppari, Å. Lernmark, J.-X. She, W. A. Hagopian, J. P. Krischer, K. Vehik, D. A. Schatz, B. Akolkar, A.-G. Ziegler, and TEDDY Study Group. 2021. An age-related exponential decline in the risk of multiple islet autoantibody seroconversion during childhood. Diabetes Care 44(10):2260–2268. https://doi.org/10.2337/dc20-2122.
Bossini-Castillo, L., G. Villanueva-Martin, M. Kerick, M. Acosta-Herrera, E. Lopez-Isac, C. P. Simeon, N. Ortego-Centeno, S. Assassi, International SSc Group, Australian Scleroderma Interest Group (ASIG), PRECISESADS Clinical Consortium, PRECISESADS Flow Cytometry study group, N. Hunzelmann, A. Gabrielli, J. K. de Vries-Bouwstra, Y. Allanore, C. Fonseca, C. P. Denton, T. RDJ Radstake, M. E. Alarcón-Riquelme, L. Beretta, M. D. Mayes, and J. Martin. 2021. Genomic risk score impact on susceptibility to systemic sclerosis. Annals of the Rheumatic Diseases 80(1):118–127. http://dx.doi.org/10.1136/annrheumdis-2020-218558.
Boudigaard, S. H., V. Schlünssen, J. M. Vestergaard, K. Søndergaard, K. Torén, S. Peters, H. Kromhout, and H. A. Kolstad. 2021. Occupational exposure to respirable crystalline silica and risk of autoimmune rheumatic diseases: A nationwide cohort study. International Journal of Epidemiology 50(4):1213–1226. https://doi.org/10.1093/ije/dyaa287.
Brix, T. H., K. O. Kyvik, K. Christensen, and L. Hegedus. 2001. Evidence for a major role of heredity in Graves’ disease: A population-based study of two Danish twin cohorts. Journal of Clinical Endocrinology & Metabolism 86(2):930–934. https://doi.org/10.1210/jcem.86.2.7242.
Brown, J. M., A. J. Archer, J. C. Pfau, and A. Holian. 2003. Silica accelerated systemic autoimmune disease in lupus-prone New Zealand mixed mice. Clinical & Experimental Immunology 131(3):415–421. https://doi.org/10.1046/j.1365-2249.2003.02094.x.
Brown, L. V., E. A. Gaffney, J. Wagg, and M. C. Coles. 2018. Applications of mechanistic modelling to clinical and experimental immunology: An emerging technology to accelerate immunotherapeutic discovery and development. Clinical & Experimental Immunology 193(3):284–292. https://doi.org/10.1111/cei.13182.
Bruno, K. A., J. E. Mathews, A. L. Yang, J. A. Frisancho, A. J. Scott, H. D. Greyner, F. A. Molina, M. S. Greenaway, G. M. Cooper, A. Bucek, A. C. Morales-Lara, A. R. Hill, A. A. Mease, D. N. Di Florio, J. M. Sousou, A. C. Coronado, A. R. Stafford, and D. Fairweather. 2019. BPA alters estrogen receptor expression in the heart after viral infection activating cardiac mast cells and T cells leading to perimyocarditis and fibrosis. Frontiers in Endocrinology 10:598. https://doi.org/10.3389/fendo.2019.00598.
Buniello, A., J. A. L. MacArthur, M. Cerezo, L. W. Harris, J. Hayhurst, C. Malangone, A. McMahon, J. Morales, E. Mountjoy, E. Sollis, D. Suveges, O. Vrousgou, P. L. Whetzel, R. Amode, J. A. Guillen, H. S. Riat, S. J. Trevanion, P. Hall, H. Junkins, P. Flicek, T. Burdett, L. A. Hindorff, F. Cunningham, and H. Parkinson. 2019. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Research 47(D1):D1005–D1012. https://doi.org/10.1093/nar/gky1120.
Burbelo, P. D., M. J. Iadarola, J. M. Keller, and B. M. Warner. 2021. Autoantibodies targeting intracellular and extracellular proteins in autoimmunity. Frontiers in Immunology 12:5.
Buskiewicz, I. A., S. A. Huber, and D. Fairweather. 2016. Sex hormone receptor expression in the immune system. In Sex differences in physiology, edited by G. Neigh and M. Mitzelfelt. Cambridge, MA: Academic Press. Pp. 45–60.
Capecchi, R., I. Puxeddu, F. Pratesi, and P. Migliorini. 2020. New biomarkers in SLE: From bench to bedside. Rheumatology (Oxford) 59(Suppl 5):v12–v18. https://doi.org/10.1093/rheumatology/keaa484.
Carey, R. N., J. C. Pfau, M. J. Fritzler, J. Creaney, N. de Klerk, A. W. B. Musk, P. Franklin, N. Sodhi-Berry, F. Brims, and A. Reid. 2021. Autoantibodies and cancer among asbestos-exposed cohorts in Western Australia. Journal of Toxicology and Environmental Health, Part A 84(11):475–483. https://doi.org/10.1080/15287394.2021.1889424.
Castro-Correia, C., L. Correia-Sa, S. Norberto, C. Delerue-Matos, V. Domingues, C. Costa-Santos, M. Fontoura, and C. Calhau. 2018. Phthalates and type 1 diabetes: Is there any link? Environmental Science and Pollution Research International 25(18):17915–17919. https://doi.org/10.1007/s11356-018-1997-z.
Cavalli, G., and E. Heard. 2019. Advances in epigenetics link genetics to the environment and disease. Nature 571(7766):489–499. https://doi.org/10.1038/s41586-019-1411-0.
Chain, J. L., K. Alvarez, A. Mascaro-Blanco, S. Reim, R. Bentley, R. Hommer, P. Grant, J. F. Leckman, I. Kawikova, K. Williams, J. A. Stoner, S. E. Swedo, and M. W. Cunningham. 2020. Autoantibody biomarkers for basal ganglia encephalitis in sydenham chorea and pediatric autoimmune neuropsychiatric disorder associated with streptococcal infections. Frontiers in Psychiatry 11:564. https://doi.org/10.3389/fpsyt.2020.00564.
Chauhan, P. S., J. G. Wagner, A. D. Benninghoff, R. P. Lewandowski, O. K. Favor, K. A. Wierenga, K. N. Gilley, E. A. Ross, J. R. Harkema, and J. J. Pestka. 2021. Rapid induction of pulmonary inflammation, autoimmune gene expression, and ectopic lymphoid neogenesis following acute silica exposure in lupus-prone mice. Frontiers in Immunology 12:635138. https://doi.org/10.3389/fimmu.2021.635138.
Cho, J. H., and P. K. Gregersen. 2011. Genomics and the multifactorial nature of human autoimmune disease. The New England Journal of Medicine 365(17):1612–1623. https://doi.org/10.1056/NEJMra1100030.
Chu, X., B. Zhang, V. A. C. M. Koeken, M. K. Gupta, and Y. Li. 2021. Multi-omics approaches in immunological research. Frontiers in Immunology 12:668045. https://doi.org/10.3389/fimmu.2021.668045.
Cojocaru, M., I. M. Cojocaru, and I. Silosi. 2010. Multiple autoimmune syndrome. Maedica (Bucur) 5(2):132–134.
Conrad, K., Y. Shoenfeld, and M. J. Fritzler. 2020. Precision health: A pragmatic approach to understanding and addressing key factors in autoimmune diseases. Autoimmunity Reviews 19(5):102508. https://doi.org/10.1016/j.autrev.2020.102508.
Consiglio, C. R., N. Cotugno, F. Sardh, C. Pou, D. Amodio, L. Rodriguez, Z. Tan, S. Zicari, A. Ruggiero, G. R. Pascucci, V. Santilli, T. Campbell, Y. Bryceson, D. Eriksson, J. Wang, A. Marchesi, T. Lakshmikanth, A. Campana, A. Villani, P. Rossi, C. S. Team, N. Landegren, P. Palma, and P. Brodin. 2020. The immunology of multisystem inflammatory syndrome in children with COVID-19. Cell 183(4):968–981 e967. https://doi.org/10.1016/j.cell.2020.09.016.
Cook, I. F. 2008. Sexual dimorphism of humoral immunity with human vaccines. Vaccine 26(29–30):3551–3555. https://doi.org/10.1016/j.vaccine.2008.04.054.
Cooper, G. S., F. W. Miller, and J. P. Pandey. 1999. The role of genetic factors in autoimmune disease: Implications for environmental research. Environmental Health Perspectives 107 Suppl 5(Suppl 5):693–700. https://doi.org/10.1289/ehp.99107s5693.
Cooper, G. S., and B. C. Stroehla. 2003. The epidemiology of autoimmune diseases. Autoimmunity Reviews 2(3):119–125. https://doi.org/10.1016/s1568-9972(03)00006-5.
Cooper, G. S., K. M. Gilbert, E. L. Greidinger, J. A. James, J. C. Pfau, L. Reinlib, B. C. Richardson, and N. R. Rose. 2008. Recent advances and opportunities in research on lupus: Environmental influences and mechanisms of disease. Environmental Health Perspectives 116(6):695–702. https://doi.org/10.1289/ehp.11092.
Cooper, G. S., S. L. Makris, P. J. Nietert, and J. Jinot. 2009. Evidence of autoimmune-related effects of trichloroethylene exposure from studies in mice and humans. Environmental Health Perspectives 117(5):696–702. https://doi.org/10.1289/ehp.11782.
Coronado, M. J., K. A. Bruno, L. A. Blauwet, C. Tschöpe, M. W. Cunningham, S. Pankuweit, S. van Linthout, E.-S. Jeon, D. M. McNamara, J. Krejčí, J. Bienertová-Vašků, E. J. Douglass, E. D. Abston, A. Bucek, J. A. Frisancho, M. S. Greenaway, A. R. Hill, H.-P. Schultheiss, L. T. Cooper, Jr., and D. Fairweather. 2019. Elevated sera sst2 is associated with heart failure in men ≤50 years old with myocarditis. Journal of the American Heart Association 8(2):e008968. https://doi.org/10.1161/JAHA.118.008968.
Costenbader, K. H., and E. W. Karlson. 2006. Cigarette smoking and autoimmune disease: What can we learn from epidemiology? Lupus 15(11):737–745. https://doi.org/10.1177/0961203306069344.
Cunningham, M. W. 2019. Molecular mimicry, autoimmunity, and infection: The cross-reactive antigens of group a streptococci and their sequelae. Microbiology Spectrum 7(4). https://doi.org/10.1128/microbiolspec.GPP3-0045-2018.
Dabelea, D., E. J. Mayer-Davis, S. Saydah, G. Imperatore, B. Linder, J. Divers, R. Bell, A. Badaru, J. W. Talton, T. Crume, A. D. Liese, A. T. Merchant, J. M. Lawrence, K. Reynolds, L. Dolan, L. L. Liu, R. F. Hamman, and Search for Diabetes in Youth Study. 2014. Prevalence of type 1 and type 2 diabetes among children and adolescents from 2001 to 2009. JAMA 311(17):1778–1786. https://doi.org/10.1001/jama.2014.3201.
Daha, N. A., N. K. Banda, A. Roos, F. J. Beurskens, J. M. Bakker, M. R. Daha, and L. A. Trouw. 2011. Complement activation by (auto-) antibodies. Molecular Immunology 48(14):1656–1665. https://doi.org/10.1016/j.molimm.2011.04.024.
De Decker, E., M. Vanthuyne, D. Blockmans, F. Houssiau, J. Lenaerts, R. Westhovens, B. Nemery, and E. De Langhe. 2018. High prevalence of occupational exposure to solvents or silica in male systemic sclerosis patients: A Belgian cohort analysis. Clinical Rheumatology 37(7):1977–1982. https://doi.org/10.1007/s10067-018-4045-y.
de Lange, K. M., L. Moutsianas, J. C. Lee, C. A. Lamb, Y. Luo, N. A. Kennedy, L. Jostins, D. L. Rice, J. Gutierrez-Achury, S. G. Ji, G. Heap, E. R. Nimmo, C. Edwards, P. Henderson, C. Mowat, J. Sanderson, J. Satsangi, A. Simmons, D. C. Wilson, M. Tremelling, A. Hart, C. G. Mathew, W. G. Newman, M. Parkes, C. W. Lees, H. Uhlig, C. Hawkey, N. J. Prescott, T. Ahmad, J. C. Mansfield, C. A. Anderson, and J. C. Barrett. 2017. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nature Genetics 49(2):256–261. https://doi.org/10.1038/ng.3760.
de Loizaga, S. R., and A. Z. Beaton. 2021. Rheumatic fever and rheumatic heart disease in the United States. Pediatric Annals 50(3):e98–e104. https://doi.org/10.3928/19382359-20210221-01.
de Mol, C. L., P. R. Jansen, R. L. Muetzel, M. J. Knol, H. H. Adams, V. W. Jaddoe, M. W. Vernooij, R. Q. Hintzen, T. J. White, and R. F. Neuteboom. 2020. Polygenic multiple sclerosis risk and population-based childhood brain imaging. Annals of Neurology 87(5):774–787. https://doi.org/10.1002/ana.25717.
Deane, K. D., and V. M. Holers. 2021. Rheumatoid arthritis pathogenesis, prediction, and prevention: An emerging paradigm shift. Arthritis & Rheumatology 73(2):181–193. https://doi.org/10.1002/art.41417.
DeBord, D. G., T. Carreon, T. J. Lentz, P. J. Middendorf, M. D. Hoover, and P. A. Schulte. 2016. Use of the “exposome” in the practice of epidemiology: A primer on -omic technologies. American Journal of Epidemiology 184(4):302–314. https://doi.org/10.1093/aje/kwv325.
Dendrou, C. A., A. Cortes, L. Shipman, H. G. Evans, K. E. Attfield, L. Jostins, T. Barber, G. Kaur, S. B. Kuttikkatte, O. A. Leach, C. Desel, S. L. Faergeman, J. Cheeseman, M. J. Neville, S. Sawcer, A. Compston, A. R. Johnson, C. Everett, J. I. Bell, F. Karpe, M. Ultsch, C. Eigenbrot, G. McVean, and L. Fugger. 2016. Resolving TYK2 locus genotype-to-phenotype differences in autoimmunity. Science Translational Medicine 8(363):363ra149. https://dx.doi.org/10.1126/scitranslmed.aag1974.
Desai, M. K., and R. D. Brinton. 2019. Autoimmune disease in women: Endocrine transition and risk across the lifespan. Frontiers in Endocrinology 10:265. https://doi.org/10.3389/fendo.2019.00265.
Di Giuseppe, D., A. Wallin, M. Bottai, J. Askling, and A. Wolk. 2014. Long-term intake of dietary long-chain n-3 polyunsaturated fatty acids and risk of rheumatoid arthritis: A prospective cohort study of women. Annals of the Rheumatic Diseases 73(11):1949–1953. https://doi.org/10.1136/annrheumdis-2013-203338.
Diamanti-Kandarakis, E., J.-P. Bourguignon, L. C. Giudice, R. Hauser, G. S. Prins, A. M. Soto, R. T. Zoeller, and A. C. Gore. 2009. Endocrine-disrupting chemicals: An endocrine society scientific statement. Endocrine Reviews 30(4):293–342. https://doi.org/10.1210/er.2009-0002.
Diegel, R., B. Black, J. C. Pfau, T. McNew, C. Noonan, and R. Flores. 2018. Case series: Rheumatological manifestations attributed to exposure to libby asbestiform amphiboles. Journal of Toxicology and Environmental Health, Part A 81(15):734–747.
Dotan, A., S. Muller, D. Kanduc, P. David, G. Halpert, and Y. Shoenfeld. 2021. The SARSCoV-2 as an instrumental trigger of autoimmunity. Autoimmunity Reviews 20(4):102792. https://doi.org/10.1016/j.autrev.2021.102792.
Dube, S. R., D. Fairweather, W. S. Pearson, V. J. Felitti, R. F. Anda, and J. B. Croft. 2009. Cumulative childhood stress and autoimmune diseases in adults. Psychosomatic Medicine 71(2):243–250. https://doi.org/10.1097/PSY.0b013e3181907888.
Dubois, P. C. A., G. Trynka, L. Franke, K. A. Hunt, J. Romanos, A. Curtotti, A. Zhernakova, G. A. Heap, R. Ádány, A. Aromaa, M. T. Bardella, L. H. van den Berg, N. A. Bockett, E. G. de la Concha, B. Dema, R. S. N. Fehrmann, M. Fernández-Arquero, S. Fiatal, E. Grandone, P. M. Green, H. J. M. Groen, R. Gwilliam, R. H. J. Houwen, S. E. Hunt, K. Kaukinen, D. Kelleher, I. Korponay-Szabo, K. Kurppa, P. MacMathuna, M. Mäki, M. C. Mazzilli, O. T. McCann, M. L. Mearin, C. A. Mein, M. M. Mirza, V. Mistry, B. Mora, K. I. Morley, C. J. Mulder, J. A. Murray, C. Nunez, E. Oosterom, R. A. Ophoff, I. Polanco, L. Peltonen, M. Platteel, A. Rybak, V. Salomaa, J. J. Schweizer, M. P. Sperandeo, G. J. Tack, G. Turner, J. H. Veldink, W. H. Verbeek, R. K. Weersma, V. M. Wolters, E. Urcelay, B. Cukrowska, L. Greco, S. L. Neuhausen, R. McManus, D. Barisani, P. Deloukas, J. C. Barrett, P. Saavalainen, C. Wijmenga, and D. A. van Heel. 2010. Multiple common variants for celiac disease influencing immune gene expression. Nature Genetics 42(4):295–302. https://doi.org/10.1038/ng.543.
Edwards, M., R. Dai, and S. A. Ahmed. 2018. Our environment shapes us: The importance of environment and sex differences in regulation of autoantibody production. Frontiers in Immunology 9:478. https://doi.org/10.3389/fimmu.2018.00478.
Ellinghaus, D., L. Jostins, S. L. Spain, A. Cortes, J. Bethune, B. Han, Y. R. Park, S. Raychaudhuri, J. G. Pouget, M. Hübenthal, T. Folseraas, Y. Wang, T. Esko, A. Metspalu, H.-J. Westra, L. Franke, T. H. Pers, R. K. Weersma, V. Collij, M. D’Amato, J. Halfvarson, A. Boeck Jensen, W. Lieb, F. Degenhardt, A. J. Forstner, A. Hofmann, The International IBD Genetics Consortium (IIBDGC), International Genetics of Ankylosing Spondylitis Consortium (IGAS), International PSC Study Group (IPSCSG), Genetic Analysis of Psoriasis Consortium (GAPC), Psoriasis Association Genetics Extension (PAGE), S. Schreiber, U. Mrowietz, B. D. Juran, K. N. Lazaridis, S. Brunak, A. M. Dale, R. C. Trembath, S. Weidinger, M. Weichenthal, E. Ellinghaus, J. T. Elder, J. N. Barker, O. A. Andreassen, D. P. McGovern, T. H. Karlsen, J. C. Barrett, M. Parkes, M. A. Brown, and A. Franke. 2016. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nature Genetics 48(5):510–518. https://doi.org/10.1038/ng.3528.
EPA (Environmental Protection Agency). 2021. Ace biomonitoring. https://www.epa.gov/americaschildrenenvironment/ace-biomonitoring (accessed July 21, 2021).
Eyre, S., J. Bowes, D. Diogo, A. Lee, A. Barton, P. Martin, A. Zhernakova, E. Stahl, S. Viatte, K. McAllister, C. I. Amos, L. Padyukov, R. E. M. Toes, T. W. J. Huizinga, C. Wijmenga, G. Trynka, L. Franke, H. J. Westra, L. Alfredsson, X. Hu, C. Sandor, P. I. W. de Bakker, S. Davila, C. C. Khor, K. K. Heng, R. Andrews, S. Edkins, S. E. Hunt, C. Langford, D. Symmons, Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate, Wellcome Trust Case Control Consortium, P. Concannon, S. Onengut-Gumuscu, S. S. Rich, P. Deloukas, M. A. Gonzalez-Gay, L. Rodriguez-Rodriguez, L. Ärlsetig, J. Martin, S. Rantapää-Dahlqvist, R. M. Plenge, S. Raychaudhuri, L. Klareskog, P. K. Gregersen, and J. Worthington. 2012. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nature Genetics 44(12):1336–1340. https://doi.org/10.1038/ng.2462.
Fairweather, D., and N. R. Rose. 2006. Immunopathogenesis of autoimmune disease. In Immunotoxicology and immunopharmacology. 3rd ed., edited by R. V. House, R. Luebke, and I. Kimber. Boca Raton, FL: CRC Press. Pp. 423–436.
Fairweather, D., Z. Kaya, G. R. Shellam, C. M. Lawson, and N. R. Rose. 2001. From infection to autoimmunity. Journal of Autoimmunity 16(3):175–186. https://doi.org/10.1006/jaut.2000.0492.
Fairweather, D., S. Yusung, S. Frisancho, M. Barrett, S. Gatewood, R. Steele, and N. R. Rose. 2003. IL-12 receptor β1 and Toll-like receptor 4 increase Il-1β- and IL-18-associated myocarditis and coxsackievirus replication. The Journal of Immunology 170(9):4731–4737. https://doi.org/10.4049/jimmunol.170.9.4731.
Fairweather, D., S. Frisancho-Kiss, and N. R. Rose. 2008. Sex differences in autoimmune disease from a pathological perspective. The American Journal of Pathology 173(3):600–609.
Fairweather, D., M. A. Petri, M. J. Coronado, and L. T. Cooper. 2012. Autoimmune heart disease: Role of sex hormones and autoantibodies in disease pathogenesis. Expert Review of Clinical Immunology 8(3):269–284. https://doi.org/10.1586/eci.12.10.
Farh, K. K.-H., A. Marson, J. Zhu, M. Kleinewietfeld, W. J. Housley, S. Beik, N. Shoresh, H. Whitton, R. J. H. Ryan, A. A. Shishkin, M. Hatan, M. J. Carrasco-Alfonso, D. Mayer, C. J. Luckey, N. A. Patsopoulos, P. L. De Jager, V. K. Kuchroo, C. B. Epstein, M. J. Daly, D. A. Hafler, and B. E. Bernstein. 2015. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518(7539):337–343. https://doi.org/10.1038/nature13835.
FDA (Food and Drug Administration). 2021. Information on Tumor Necrosis Factor (TNF) blockers (marketed as Remicade, Enbrel, Humira, Cimzia, and Simponi). https://www.fda.gov/drugs/postmarket-drug-safety-information-patients-and-providers/information-tumor-necrosis-factor-tnf-blockers-marketed-remicade-enbrel-humira-cimzia-and-simponi (accessed March 22, 2022).
Fechtner, S., H. Berens, E. Bemis, R. L. Johnson, C. J. Guthridge, N. E. Carlson, M. K. Demoruelle, J. B. Harley, J. D. Edison, J. A. Norris, W. H. Robinson, K. D. Deane, J. A. James, and V. M. Holers. 2021. Antibody responses to Epstein-Barr virus in the preclinical period of rheumatoid arthritis suggest the presence of increased viral reactivation cycles. Arthritis & Rheumatology. https://doi.org/10.1002/art.41994.
Feghali-Bostwick, C., T. A. Medsger, Jr., and T. M. Wright. 2003. Analysis of systemic sclerosis in twins reveals low concordance for disease and high concordance for the presence of antinuclear antibodies. Arthritis and Rheumatism 48(7):1956–1963. https://doi.org/10.1002/art.11173.
Feldmann, M., and R. N. Maini. 2010. Anti-TNF therapy, from rationale to standard of care: What lessons has it taught us? The Journal of Immunology 185(2):791–794. https://doi.org/10.4049/jimmunol.1090051.
Ferro, A., C. N. Zebedeo, C. Davis, K. W. Ng, and J. C. Pfau. 2014. Amphibole, but not chryso-tile, asbestos induces anti-nuclear autoantibodies and IL-17 in C57BL/6 mice. Journal of Immunotoxicology 11(3):283–290. https://doi.org/10.3109/1547691x.2013.847510.
Fischinger, S., C. M. Boudreau, A. L. Butler, H. Streeck, and G. Alter. 2019. Sex differences in vaccine-induced humoral immunity. Seminars in Immunopathology 41(2):239–249. https://doi.org/10.1007/s00281-018-0726-5.
Flanagan, K. L., A. L. Fink, M. Plebanski, and S. L. Klein. 2017. Sex and gender differences in the outcomes of vaccination over the life course. Annual Review of Cell and Developmental Biology 33:577–599. https://doi.org/10.1146/annurev-cellbio-100616-060718.
Fontes, J. A., J. G. Barin, M. V. Talor, N. Stickel, J. Schaub, N. R. Rose, and D. Čiháková. 2017. Complete Freund’s adjuvant induces experimental autoimmune myocarditis by enhancing IL-6 production during initiation of the immune response. Immunity, Inflammation and Disease 5(2):163–176. https://doi.org/10.1002/iid3.155.
Foster, M. H., J. R. Ord, E. J. Zhao, A. Birukova, L. Fee, F. M. Korte, Y. G. Asfaw, V. L. Roggli, A. J. Ghio, R. M. Tighe, and A. G. Clark. 2019. Silica exposure differentially modulates autoimmunity in lupus strains and autoantibody transgenic mice. Frontiers in Immunology 10:2336. https://doi.org/10.3389/fimmu.2019.02336.
Frederiksen, B., M. Kroehl, M. M. Lamb, J. Seifert, K. Barriga, G. S. Eisenbarth, M. Rewers, and J. M. Norris. 2013. Infant exposures and development of type 1 diabetes mellitus: The diabetes autoimmunity study in the young (DAISY). JAMA Pediatrics 167(9):808–815. https://doi.org/10.1001/jamapediatrics.2013.317.
Gan, R. W., E. A. Bemis, M. K. Demoruelle, C. C. Striebich, S. Brake, M. L. Feser, L. Moss, M. Clare-Salzler, V. M. Holers, K. D. Deane, and J. M. Norris. 2017a. The association between omega-3 fatty acid biomarkers and inflammatory arthritis in an anti-citrullinated protein antibody positive population. Rheumatology (Oxford) 56(12):2229–2236. https://doi.org/10.1093/rheumatology/kex360.
Gan, R. W., M. K. Demoruelle, K. D. Deane, M. H. Weisman, J. H. Buckner, P. K. Gregersen, T. R. Mikuls, J. R. O’Dell, R. M. Keating, T. E. Fingerlin, G. O. Zerbe, M. J. Clare-Salzler, V. M. Holers, and J. M. Norris. 2017b. Omega-3 fatty acids are associated with a lower prevalence of autoantibodies in shared epitope-positive subjects at risk for rheumatoid arthritis. Annals of the Rheumatic Diseases 76(1):147–152. https://doi.org/10.1136/annrheumdis-2016-209154.
Gan, R. W., K. A. Young, G. O. Zerbe, M. K. Demoruelle, M. H. Weisman, J. H. Buckner, P. K. Gregersen, T. R. Mikuls, J. R. O’Dell, R. M. Keating, M. J. Clare-Salzler, K. D. Deane, V. M. Holers, and J. M. Norris. 2016. Lower omega-3 fatty acids are associated with the presence of anti-cyclic citrullinated peptide autoantibodies in a population at risk for future rheumatoid arthritis: A nested case-control study. Rheumatology (Oxford) 55(2):367–376. https://doi.org/10.1093/rheumatology/kev266.
George, A., T. Diana, J. Langericht, and G. J. Kahaly. 2021. Stimulatory thyrotropin receptor antibodies are a biomarker for Graves’ orbitopathy. Frontiers in Endocrinology 11:629925. https://doi.org/10.3389/fendo.2020.629925.
Giorgiutti, S., Y. Dieudonné, O. Hinschberger, B. Nespola, J. Campagne, H. N. Rakotoarivelo, T. Hannedouche, B. Moulin, G. Blaison, J.-C. Weber, M.-C. Dalmas, F. De Blay, D. Lipsker, F. Chantrel, J.-E. Gottenberg, Y. Dimitrov, O. Imhoff, P.-E. Gavand, E. Andres, C. Debry, Y. Hansmann, A. Klein, C. Lohmann, F. Mathiaux, A. Guffroy, V. Poindron, T. Martin, A.-S. Korganow, and L. Arnaud. 2021. Prevalence of antineutrophil cytoplasmic antibody-associated vasculitis and spatial association with quarries in a region of northeastern France: A capture-recapture and geospatial analysis. Arthritis & Rheumatology 73(11):2078–2085. https://doi.org/10.1002/art.41767.
Gómez-Puerta, J. A., L. Gedmintas, and K. H. Costenbader. 2013. The association between silica exposure and development of ANCA-associated vasculitis: Systematic review and meta-analysis. Autoimmunity Reviews 12(12):1129–1135. https://doi.org/10.1016/j.autrev.2013.06.016.
Gonzalez-Quintial, R., J. M. Mayeux, D. H. Kono, A. N. Theofilopoulos, K. M. Pollard, and R. Baccala. 2019. Silica exposure and chronic virus infection synergistically promote lupus-like systemic autoimmunity in mice with low genetic predisposition. Clinical Immunology 205:75–82. https://doi.org/10.1016/j.clim.2019.06.003.
Gubbels Bupp, M. R., and T. N. Jorgensen. 2018. Androgen-induced immunosuppression. Frontiers in Immunology 9:794. https://doi.org/10.3389/fimmu.2018.00794.
Gulamhusein, A. F., B. D. Juran, and K. N. Lazaridis. 2015. Genome-wide association studies in primary biliary cirrhosis. Seminars in Liver Disease 35(4):392–401. https://doi.org/10.1055/s-0035-1567831.
Hahn, J., N. R. Cook, E. K. Alexander, S. Friedman, J. Walter, V. Bubes, G. Kotler, I. M. Lee, J. E. Manson, and K. H. Costenbader. 2022. Vitamin D and marine omega 3 fatty acid supplementation and incident autoimmune disease: Vital randomized controlled trial. BMJ 376:e066452. https://doi.org/10.1136/bmj-2021-066452.
Hale, W. G., J. P. Margham, and V. A. Saunders, editors. 2005. S.V. “Self-tolerance.” In Collins Dictionary of Biology. New York, NY: Harper Collins.
Hamilton-Williams, E. E., G. L. Lorca, J. M. Norris, and J. L. Dunne. 2021. A triple threat? The role of diet, nutrition, and the microbiota in T1D pathogenesis. Frontiers in Nutrition 8:600756. https://doi.org/10.3389/fnut.2021.600756.
Harrington, R., S. A. Al Nokhatha, and R. Conway. 2020. JAK inhibitors in rheumatoid arthritis: An evidence-based review on the emerging clinical data. Journal of Inflammation Research 13:519–531. https://doi.org/10.2147/JIR.S219586.
Hassanalilou, T., L. Khalili, S. Ghavamzadeh, A. Shokri, L. Payahoo, and Y. K. Bishak. 2018. Role of vitamin D deficiency in systemic lupus erythematosus incidence and aggravation. Autoimmunity Highlights 9(1):1. https://doi.org/10.1007/s13317-017-0101-x.
Hedström, A. K., J. Hillert, T. Olsson, and L. Alfredsson. 2021. Factors affecting the risk of relapsing-onset and progressive-onset multiple sclerosis. Journal of Neurology, Neurosurgery & Psychiatry 92:1096–1102. http://dx.doi.org/10.1136/jnnp-2020-325688.
Herold, K. C., B. N. Bundy, S. A. Long, J. A. Bluestone, L. A. DiMeglio, M. J. Dufort, S. E. Gitelman, P. A. Gottlieb, J. P. Krischer, P. S. Linsley, J. B. Marks, W. Moore, A. Moran, H. Rodriguez, W. E. Russell, D. Schatz, J. S. Skyler, E. Tsalikian, D. K. Wherrett, A.-G. Ziegler, C. J. Greenbaum, for the Type 1 Diabetes TrialNet Study Group. 2019. An antiCD3 antibody, teplizumab, in relatives at risk for type 1 diabetes. The New England Journal of Medicine 381(7):603–613. https://doi.org/10.1056/nejmoa1902226.
Hou, J. K., B. Abraham, and H. El-Serag. 2011. Dietary intake and risk of developing inflammatory bowel disease: A systematic review of the literature. The American Journal of Gastroenterology 106(4):563–573. https://doi.org/10.1038/ajg.2011.44.
Hu, Y., K. H. Costenbader, X. Gao, M. Al-Daabil, J. A. Sparks, D. H. Solomon, F. B. Hu, E. W. Karlson, and B. Lu. 2014. Sugar-sweetened soda consumption and risk of developing rheumatoid arthritis in women. American Journal of Clinical Nutrition 100(3):959–967. https://doi.org/10.3945/ajcn.114.086918.
Idborg, H., and V. Oke. 2021. Cytokines as biomarkers in systemic lupus erythematosus: Value for diagnosis and drug therapy. International Journal of Molecular Sciences 22(21). https://doi.org/10.3390/ijms222111327.
Iezzoni, L. I. 2013. Risk adjustment for measuring health care outcomes, 4th ed. Arlington, VA: Chicago, IL: Health Administration Press.
Ilar, A., L. Klareskog, S. Saevarsdottir, P. Wiebert, J. Askling, P. Gustavsson, and L. Alfredsson. 2019. Occupational exposure to asbestos and silica and risk of developing rheumatoid arthritis: Findings from a Swedish population-based case-control study. RMD Open 5(2):e000978. https://doi.org/10.1136/rmdopen-2019-000978.
Imgenberg-Kreuz, J., A. Rasmussen, K. Sivils, and G. Nordmark. 2021. Genetics and epigenetics in primary Sjögren’s syndrome. Rheumatology (Oxford) 60(5):2085–2098. https://doi.org/10.1093/rheumatology/key330.
International Genetics of Ankylosing Spondylitis Consortium (IGAS), A. Cortes, J. Hadler, J. P. Pointon, P. C. Robinson, T. Karaderi, P. Leo, K. Cremin, K. Pryce, J. Harris, S. Lee, K. B. Joo, S.-C. Shim, M. Weisman, M. Ward, X. Zhou, H.-J. Garchon, G. Chiocchia, J. Nossent, B. A. Lie, O. Førre, J. Tuomilehto, K. Laiho, L. Jiang, Y. Liu, X. Wu, L. A. Bradbury, D. Elewaut, R. Burgos-Vargas, S. Stebbings, L. Appleton, C. Farrah, J. Lau, T. J. Kenna, N. Haroon, M. A. Ferreira, J. Yang, J. Mulero, J. L. Fernandez-Sueiro, M. A. Gonzalez-Gay, C. Lopez-Larrea, P. Deloukas, P. Donnelly, Australo-Anglo-American Spondyloarthritis Consortium, Groupe Francaise d’Etude Genetique des Spondylarthrites, NordTrondelag Health Study, Spondyloarthritis Research Consortium of Canada, Wellcome Trust Case Control Consortium, P. Bowness, K. Gafney, H. Gaston, D. D. Gladman, P. Rahman, W. P. Maksymowych, H. Xu, J. B. Crusius, I. E. van der Horst-Bruinsma, C. T. Chou, R. Valle-Onate, C. Romero-Sanchez, I. M. Hansen, F. M. Pimentel-Santos, R. D. Inman, V. Videm, J. Martin, M. Breban, J. D. Reveille, D. M. Evans, T. H. Kim, B. P. Wordsworth, and M. A. Brown. 2013. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nature Genetics 45(7):730–738. https://doi.org/10.1038/ng.2667.
International Multiple Sclerosis Genetics Consortium (IMSGC). 2019. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365(6460). https://doi.org/10.1126/science.aav7188.
International Multiple Sclerosis Genetics Consortium (IMSGC), A. H. Beecham, N. A. Patsopoulos, D. K. Xifara, M. F. Davis, A. Kemppinen, C. Cotsapas, T. S. Shah, C. Spencer, D. Booth, A. Goris, A. Oturai, J. Saarela, B. Fontaine, B. Hemmer, C. Martin, F. Zipp, S. D’Alfonso, F. Martinelli-Boneschi, B. Taylor, H. F. Harbo, I. Kockum, J. Hillert, T. Olsson, M. Ban, J. R. Oksenberg, R. Hintzen, L. F. Barcellos, Wellcome Trust Case Control Consortium 2 (WTCCC2), International IBD Genetics Consortium (IIBDGC), C. Agliardi, L. Alfredsson, M. Alizadeh, C. Anderson, R. Andrews, H. B. Søndergaard, A. Baker, G. Band, S. E. Baranzini, N. Barizzone, J. Barrett, C. Bellenguez, L. Bergamaschi, L. Bernardinelli, A. Berthele, V. Biberacher, T. M. Binder, H. Blackburn, I. L. Bomfim, P. Brambilla, S. Broadley, B. Brochet, L. Brundin, D. Buck, H. Butzkueven, S. J. Caillier, W. Camu, W. Carpentier, P. Cavalla, E. G. Celius, I. Coman, G. Comi, L. Corrado, L. Cosemans, I. Cournu-Rebeix, B. A. Cree, D. Cusi, V. Damotte, G. Defer, S. R. Delgado, P. Deloukas, A. di Sapio, A. T. Dilthey, P. Donnelly, B. Dubois, M. Duddy, S. Edkins, I. Elovaara, F. Esposito, N. Evangelou, B. Fiddes, J. Field, A. Franke, C. Freeman, I. Y. Frohlich, D. Galimberti, C. Gieger, P. A. Gourraud, C. Graetz, A. Graham, V. Grummel, C. Guaschino, A. Hadjixenofontos, H. Hakonarson, C. Halfpenny, G. Hall, P. Hall, A. Hamsten, J. Harley, T. Harrower, C. Hawkins, G. Hellenthal, C. Hillier, J. Hobart, M. Hoshi, S. E. Hunt, M. Jagodic, I. Jelčić, A. Jochim, B. Kendall, A. Kermode, T. Kilpatrick, K. Koivisto, I. Konidari, T. Korn, H. Kronsbein, C. Langford, M. Larsson, M. Lathrop, C. Lebrun-Frenay, J. Lechner-Scott, M. H. Lee, M. A. Leone, V. Leppä, G. Liberatore, B. A. Lie, C. M. Lill, M. Lindén, J. Link, F. Luessi, J. Lycke, F. Macciardi, S. Männistö, C. P. Manrique, R. Martin, V. Martinelli, D. Mason, G. Mazibrada, C. McCabe, I. L. Mero, J. Mescheriakova, L. Moutsianas, K. M. Myhr, G. Nagels, R. Nicholas, P. Nilsson, F. Piehl, M. Pirinen, S. E. Price, H. Quach, M. Reunanen, W. Robberecht, N. P. Robertson, M. Rodegher, D. Rog, M. Salvetti, N. C. Schnetz-Boutaud, F. Sellebjerg, R. C. Selter, C. Schaefer, S. Shaunak, L. Shen, S. Shields, V. Siffrin, M. Slee, P. S. Sorensen, M. Sorosina, M. Sospedra, A. Spurkland, A. Strange, E. Sundqvist, V. Thijs, J. Thorpe, A. Ticca, P. Tienari, C. van Duijn, E. M. Visser, S. Vucic, H. Westerlind, J. S. Wiley, A. Wilkins, J. F. Wilson, J. Winkelmann, J. Zajicek, E. Zindler, J. L. Haines, M. A. Pericak-Vance, A. J. Ivinson, G. Stewart, D. Hafler, S. L. Hauser, A. Compston, G. McVean, P. De Jager, S. J. Sawcer, and J. L. McCauley. 2013. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nature Genetics 45(11):1353–1360. https://doi.org/10.1038/ng.2770.
Izmirly, P. M., H. Parton, L. Wang, W. J. McCune, S. S. Lim, C. Drenkard, E. D. Ferucci, M. Dall’Era, C. Gordon, C. G. Helmick, and E. C. Somers. 2021. Prevalence of systemic lupus erythematosus in the United States: Estimates from a meta-analysis of the centers for disease control and prevention national lupus registries. Arthritis & Rheumatology:1–6. https://doi.org/10.1002/art.41632.
Jacobsen, R., E. Hypponen, T. I. Sorensen, A. A. Vaag, and B. L. Heitmann. 2015. Gestational and early infancy exposure to margarine fortified with vitamin D through a national Danish programme and the risk of type 1 diabetes: The D-Tect study. PloS One 10(6):e0128631. https://doi.org/10.1371/journal.pone.0128631.
James, J. A., H. Chen, K. A. Young, E. A. Bemis, J. Seifert, R. L. Bourn, K. D. Deane, M. K. Demoruelle, M. Feser, J. R. O’Dell, M. H. Weisman, R. M. Keating, P. M. Gaffney, J. A. Kelly, C. D. Langefeld, J. B. Harley, W. Robinson, D. A. Hafler, K. C. O’Connor, J. Buckner, J. M. Guthridge, J. M. Norris, and V. M. Holers. 2019. Latent autoimmunity across disease-specific boundaries in at-risk first-degree relatives of SLE and RA patients. EBioMedicine 42:76–85. https://doi.org/10.1016/j.ebiom.2019.03.063.
Jang, D. I., A. H. Lee, H. Y. Shin, H. R. Song, J. H. Park, T. B. Kang, S. R. Lee, and S. H. Yang. 2021. The role of tumor necrosis factor alpha (tnf-alpha) in autoimmune disease and current tnf-alpha inhibitors in therapeutics. International Journal of Molecular Sciences 22(5). https://doi.org/10.3390/ijms22052719.
Jantchou, P., S. Morois, F. Clavel-Chapelon, M.-C. Boutron-Ruault, and F. Carbonnel. 2010. Animal protein intake and risk of inflammatory bowel disease: The E3N prospective study. The American Journal of Gastroenterology 105(10):2195–2201. https://doi.org/10.1038/ajg.2010.192.
Jesus, A. A., and R. Goldbach-Mansky. 2014. IL-1 blockade in autoinflammatory syndromes. Annual Review of Medicine 65:223–244. https://doi.org/10.1146/annurevmed-061512-150641.
Jiang, J., M. Zhao, C. Chang, H. Wu, and Q. Lu. 2020. Type I interferons in the pathogenesis and treatment of autoimmune diseases. Clinical Reviews in Allergy & Immunology 59(2):248–272. https://doi.org/10.1007/s12016-020-08798-2.
Joachims, M. L., K. M. Leehan, C. Lawrence, R. C. Pelikan, J. S. Moore, Z. Pan, A. Rasmussen, L. Radfar, D. M. Lewis, K. M. Grundahl, J. A. Kelly, G. B. Wiley, M. Shugay, D. M. Chudakov, C. J. Lessard, D. U. Stone, R. H. Scofield, C. G. Montgomery, K. L. Sivils, L. F. Thompson, and A. D. Farris. 2016. Single-cell analysis of glandular T cell receptors in Sjogren’s syndrome. JCI Insight 1(8). https://doi.org/10.1172/jci.insight.85609.
Jog, N. R., K. A. Young, M. E. Munroe, M. T. Harmon, J. M. Guthridge, J. A. Kelly, D. L. Kamen, G. S. Gilkeson, M. H. Weisman, D. R. Karp, P. M. Gaffney, J. B. Harley, D. J. Wallace, J. M. Norris, and J. A. James. 2019. Association of Epstein-Barr virus serological reactivation with transitioning to systemic lupus erythematosus in at-risk individuals. Annals of the Rheumatic Diseases 78(9):1235–1241. https://doi.org/10.1136/annrheumdis-2019-215361.
Jostins, L., S. Ripke, R. K. Weersma, R. H. Duerr, D. P. McGovern, K. Y. Hui, J. C. Lee, L. P. Schumm, Y. Sharma, C. A. Anderson, J. Essers, M. Mitrovic, K. Ning, I. Cleynen, E. Theatre, S. L. Spain, S. Raychaudhuri, P. Goyette, Z. Wei, C. Abraham, J.-P. Achkar, T. Ahmad, L. Amininejad, A. N. Ananthakrishnan, V. Andersen, J. M. Andrews, L. Baidoo, T. Balschun, P. A. Bampton, A. Bitton, G. Boucher, S. Brand, C. Büning, A. Cohain, S. Cichon, M. D’Amato, D. De Jong, K. L. Devaney, M. Dubinsky, C. Edwards, D. Ellinghaus, L. R. Ferguson, D. Franchimont, K. Fransen, R. Gearry, M. Georges, C. Gieger, J. Glas, T. Haritunians, A. Hart, C. Hawkey, M. Hedl, X. Hu, T. H. Karlsen, L. Kupcinskas, S. Kugathasan, A. Latiano, D. Laukens, I. C. Lawrance, C. W. Lees, E. Louis, G. Mahy, J. Mansfield, A. R. Morgan, C. Mowat, W. Newman, O. Palmieri, C. Y. Ponsioen, U. Potocnik, N. J. Prescott, M. Regueiro, J. I. Rotter, R. K. Russell, J. D. Sanderson, M. Sans, J. Satsangi, S. Schreiber, L. A. Simms, J. Sventoraityte, S. R. Targan, K. D. Taylor, M. Tremelling, H. W. Verspaget, M. De Vos, C. Wijmenga, D. C. Wilson, J. Winkelmann, R. J. Xavier, S. Zeissig, B. Zhang, C. K. Zhang, H. Zhao, The International IBD Genetics Consortium (IIBDGC), M. S. Silverberg, V. Annese, H. Hakonarson, S. R. Brant, G. Radford-Smith, C. G. Mathew, J. D. Rioux, E. E. Schadt, M. J. Daly, A. Franke, M. Parkes, S. Vermeire, J. C. Barrett, and J. H. Cho. 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491(7422):119–124. https://doi.org/10.1038/nature11582.
Kawakami, R., A. Sakamoto, K. Kawai, A. Gianatti, D. Pellegrini, A. Nasr, B. Kutys, L. Guo, A. Cornelissen, M. Mori, Y. Sato, I. Pescetelli, M. Brivio, M. Romero, G. Guagliumi, R. Virmani, and A. V. Finn. 2021. Pathological evidence for SARS-CoV-2 as a cause of myocarditis: JACC review topic of the week. Journal of the American College of Cardiology 77(3):314–325. https://doi.org/10.1016/j.jacc.2020.11.031.
Kemanetzoglou, E., and E. Andreadou. 2017. CNS demyelination with TNF-alpha blockers. Current Neurology and Neuroscience Reports 17(4):36. https://doi.org/10.1007/s11910-017-0742-1.
Khaliullin, T. O., E. R. Kisin, S. Guppi, N. Yanamala, V. Zhernovkov, and A. A. Shvedova. 2020. Differential responses of murine alveolar macrophages to elongate mineral particles of asbestiform and non-asbestiform varieties: Cytotoxicity, cytokine secretion and transcriptional changes. Toxicology and Applied Pharmacology 409:115302. https://doi.org/10.1016/j.taap.2020.115302.
Khan, M. F., and H. Wang. 2019. Environmental exposures and autoimmune diseases: Contribution of gut microbiome. Frontiers in Immunology 10:3094. https://doi.org/10.3389/fimmu.2019.03094.
Khatri, B., T. R. Reksten, K. L. Tessneer, A. Rasmussen, R. H. Scofield, S. J. Bowman, J. Guthridge, J. A. James, L. Ronnblom, B. M. Warner, X. Mariette, R. Omdal, J. M. Ibanez, M. Teruel, J. L. Jensen, L. A. Aqrawi, Ø. Palm, M. Wahren-Herlenius, T. Witte, R. Jonsson, M. Rischmueller, A. D. Farris, M. Alarcon-Riquelme, W. F. Ng, K. L. Sivils, G. Nordmark, and C. Lessard. 2020 November 9, 2020. Genome-wide association study of Sjögren’s syndrome identifies 10 new risk loci. Paper presented at ACR Convergence 2020.
Klement, E., R. V. Cohen, J. Boxman, A. Joseph, and S. Reif. 2004. Breastfeeding and risk of inflammatory bowel disease: A systematic review with meta-analysis. American Journal of Clinical Nutrition 80(5):1342–1352. https://doi.org/10.1093/ajcn/80.5.1342.
Kochi, Y. 2016. Genetics of autoimmune diseases: Perspectives from genome-wide association studies. International Immunology 28(4):155–161. https://doi.org/10.1093/intimm/dxw002.
Kojima, Y., A. Uzawa, Y. Ozawa, M. Yasuda, Y. Onishi, H. Akamine, N. Kawaguchi, K. Himuro, Y. I. Noto, T. Mizuno, and S. Kuwabara. 2021. Rate of change in acetylcholine receptor antibody levels predicts myasthenia gravis outcome. Journal of Neurology, Neurosurgery & Psychiatry. 92(9). http://dx.doi.org/10.1136/jnnp-2020-325511.
Krischer, J. P., X. Liu, K. Vehik, B. Akolkar, W. A. Hagopian, M. J. Rewers, J. X. She, J. Toppari, A. G. Ziegler, and Å. Lernmark. 2019. Predicting islet cell autoimmunity and type 1 diabetes: An 8-year TEDDY study progress report. Diabetes Care 42(6):1051–1060. https://doi.org/10.2337/dc18-2282.
Kumagai-Takei, N., S. Lee, B. Srinivas, Y. Shimizu, N. Sada, K. Yoshitome, T. Ito, Y. Nishimura, and T. Otsuki. 2020. The effects of asbestos fibers on human T cells. International Journal of Molecular Sciences 21(19). https://doi.org/10.3390/ijms21196987.
Kuret, T., S. Sodin-Semrl, B. Leskosek, and P. Ferk. 2021. Single cell rna sequencing in autoimmune inflammatory rheumatic diseases: Current applications, challenges and a step toward precision medicine. Frontiers in Medicine (Lausanne) 8:822804. https://doi.org/10.3389/fmed.2021.822804.
Lambers, W. M., J. Westra, H. Bootsma, and K. de Leeuw. 2021. From incomplete to complete systemic lupus erythematosus; a review of the predictive serological immune markers. Seminars in Arthritis and Rheumatism 51(1):43–48. https://doi.org/10.1016/j.semarthrit.2020.11.006.
Lane, J. R., D. A. Neumann, A. Lafond-Walker, A. Herskowitz, and N. R. Rose. 1992. Interleukin 1 or tumor necrosis factor can promote Coxsackie B3-induced myocarditis in resistant B10.A mice. Journal of Experimental Medicine 175(4):1123–1129. https://doi.org/10.1084/jem.175.4.1123.
Lane, J. R., D. A. Neumann, A. Lafond-Walker, A. Herskowitz, and N. R. Rose. 1993. Role of IL-1 and tumor necrosis factor in Coxsackie virus-induced autoimmune myocarditis. The Journal of Immunology 151(3):1682–1690.
Langefeld, C. D., H. C. Ainsworth, D. S. Cunninghame Graham, J. A. Kelly, M. E. Comeau, M. C. Marion, T. D. Howard, P. S. Ramos, J. A. Croker, D. L. Morris, J. K. Sandling, J. C. Almlöf, E. M. Acevedo-Vásquez, G. S. Alarcón, A. M. Babini, V. Baca, A. A. Bengtsson, G. A. Berbotto, M. Bijl, E. E. Brown, H. I. Brunner, M. H. Cardiel, L. Catoggio, R. Cervera, J. M. Cucho-Venegas, S. R. Dahlqvist, S. D’Alfonso, B. M. Da Silva, I. de la Rúa Figueroa, A. Doria, J. C. Edberg, E. Endreffy, J. A. Esquivel-Valerio, P. R. Fortin, B. I. Freedman, J. Frostegård, M. A. García, I. G. de la Torre, G. S. Gilkeson, D. D. Gladman, I. Gunnarsson, J. M. Guthridge, J. L. Huggins, J. A. James, C. G. M. Kallenberg, D. L. Kamen, D. R. Karp, K. M. Kaufman, L. C. Kottyan, L. Kovács, H. Laustrup, B. R. Lauwerys, Q.-Z. Li, M. A. Maradiaga-Ceceña, J. Martín, J. M. McCune, D. R. McWilliams, J. T. Merrill, P. Miranda, J. F. Moctezuma, S. K. Nath, T. B. Niewold, L. Orozco, N. Ortego-Centeno, M. Petri, C. A. Pineau, B. A. Pons-Estel, J. Pope, P. Raj, R. Ramsey-Goldman, J. D. Reveille, L. P. Russell, J. M. Sabio, C. A. Aguilar-Salinas, H. R. Scherbarth, R. Scorza, M. F. Seldin, C. Sjöwall, E. Svenungsson, S. D. Thompson, S. M. A. Toloza, L. Truedsson, T. Tusié-Luna, C. Vasconcelos, L. M. Vilá, D. J. Wallace, M. H. Weisman, J. E. Wither, T. Bhangale, J. R. Oksenberg, J. D. Rioux, P. K. Gregersen, A. C. Syvänen, L. Rönnblom, L. A. Criswell, C. O. Jacob, K. L. Sivils, B. P. Tsao, L. E. Schanberg, T. W. Behrens, E. D. Silverman, M. E. Alarcón-Riquelme, R. P. Kimberly, J. B. Harley, E. K. Wakeland, R. R. Graham, P. M. Gaffney, and T. J. Vyse. 2017. Transancestral mapping and genetic load in systemic lupus erythematosus. Nature Communications 8:16021. https://doi.org/10.1038/ncomms16021.
Lanz, T. V., R. C. Brewer, P. P. Ho, J. S. Moon, K. M. Jude, D. Fernandez, R. A. Fernandes, A. M. Gomez, G. S. Nadj, C. M. Bartley, R. D. Schubert, I. A. Hawes, S. E. Vazquez, M. Iyer, J. B. Zuchero, B. Teegen, J. E. Dunn, C. B. Lock, L. B. Kipp, V. C. Cotham, B. M. Ueberheide, B. T. Aftab, M. S. Anderson, J. L. DeRisi, M. R. Wilson, R. J. M. Bashford-Rogers, M. Platten, K. C. Garcia, L. Steinman, and W. H. Robinson. 2022. Clonally expanded B cells in multiple sclerosis bind EBV EBNA1 and GlialCAM. Nature. https://doi.org/10.1038/s41586-022-04432-7.
Lazear, H. M., J. W. Schoggins, and M. S. Diamond. 2019. Shared and distinct functions of type I and type III interferons. Immunity 50(4):907–923. https://doi.org/10.1016/j.immuni.2019.03.025.
Lee, E., Y. Kim, S. Y. Kim, and D. Kang. 2020. Asbestos exposure and autoantibody titers. Annals of Occupational and Environmental Medicine 32:e32. https://doi.org/10.35371/aoem.2020.32.e32.
Leslie, D., P. Lipsky, and A. L. Notkins. 2001. Autoantibodies as predictors of disease. Journal of Clinical Investigation 108(10):1417–1422. https://doi.org/10.1172/jci14452.
Lessard, C., I. Adrianto, J. Ice, A. Rasmussen, K. Grundahl, J. Kelly, R. Scofield, S. Bowman, S. Lester, P. Eriksson, M. Eloranta, J. Brun, L. Goransson, E. Harboe, M. Kvarnström, M. Brennan, J. Chodosh, R. Gopalakrishnan, A. Huang, P. Hughes, D. Lewis, M. Rohrer, D. Stone, N. Rhodus, B. Segal, L. Radfar, A. Farris, J. Guthridge, P. Gaffney, J. James, J. Harley, L. Rönnblom, J. Anaya, D. Cunninghame-Graham, T. Vyse, I. Alevizos, X. Mariette, R. Omdal, M. Wahren-Herlenius, T. Witte, R. Jonsson, M. Rischmueller, L. Criswell, C. Montgomery, W. Ng, G. Nordmark, and K. Sivils. 2016 September 28, 2016. Genome-wide association study identifies novel Sjögren’s syndrome risk loci in the regions of nab1, tyk2, and pttg1-mir146a. Paper presented at 2016 ACR/ARHP Annual Meeting.
Li, Y. R., J. Li, S. D. Zhao, J. P. Bradfield, F. D. Mentch, S. M. Maggadottir, C. Hou, D. J. Abrams, D. Chang, F. Gao, Y. Guo, Z. Wei, J. J. Connolly, C. J. Cardinale, M. Bakay, J. T. Glessner, D. Li, C. Kao, K. A. Thomas, H. Qiu, R. M. Chiavacci, C. E. Kim, F. Wang, J. Snyder, M. D. Richie, B. Flatø, O. Førre, L. A. Denson, S. D. Thompson, M. L. Becker, S. L. Guthery, A. Latiano, E. Perez, E. Resnick, R. K. Russell, D. C. Wilson, M. S. Silverberg, V. Annese, B. A. Lie, M. Punaro, M. C. Dubinsky, D. S. Monos, C. Strisciuglio, A. Staiano, E. Miele, S. Kugathasan, J. A. Ellis, J. E. Munro, K. E. Sullivan, C. A. Wise, H. Chapel, C. Cunningham-Rundles, S. F. Grant, J. S. Orange, P. M. Sleiman, E. M. Behrens, A. M. Griffiths, J. Satsangi, T. H. Finkel, A. Keinan, E. T. Prak, C. Polychronakos, R. N. Baldassano, H. Li, B. J. Keating, and H. Hakonarson. 2015. Meta-analysis of shared genetic architecture across ten pediatric autoimmune diseases. Nature Medicine 21(9):1018–1027. https://doi.org/10.1038/nm.3933.
Liang, B., C. Ge, E. Lonnblom, X. Lin, H. Feng, L. Xiao, J. Bai, B. Ayoglu, P. Nilsson, K. S. Nandakumar, M. Zhao, and R. Holmdahl. 2019. The autoantibody response to cyclic citrullinated collagen type ii peptides in rheumatoid arthritis. Rheumatology (Oxford, England) 58(9):1623–1633. https://doi.org/10.1093/rheumatology/kez073.
Liu, X., S. K. Tedeschi, M. Barbhaiya, C. L. Leatherwood, C. B. Speyer, B. Lu, K. H. Costenbader, E. W. Karlson, and J. A. Sparks. 2019. Impact and timing of smoking cessation on reducing risk for rheumatoid arthritis among women in the Nurses’ Health studies. Arthritis Care & Research 71(7):914–924. https://doi.org/10.1002/acr.23837.
Lund-Blix, N. A., S. Dydensborg Sander, K. Stordal, A. M. Nybo Andersen, K. S. Ronningen, G. Joner, T. Skrivarhaug, P. R. Njolstad, S. Husby, and L. C. Stene. 2017. Infant feeding and risk of type 1 diabetes in two large Scandinavian birth cohorts. Diabetes Care 40(7):920–927. https://doi.org/10.2337/dc17-0016.
Makin, S. 2021. Cracking the code of autoimmunity. Nature 595.
Malcova, H., T. Milota, Z. Strizova, D. Cebecauerova, I. Striz, A. Sediva, and R. Horvath. 2021. Interleukin-1 blockade in polygenic autoinflammatory disorders: Where are we now? Frontiers in Pharmacology 11:619273.
Manrai, A. K., Y. Cui, P. R. Bushel, M. Hall, S. Karakitsios, C. J. Mattingly, M. Ritchie, C. Schmitt, D. A. Sarigiannis, D. C. Thomas, D. Wishart, D. M. Balshaw, and C. J. Patel. 2017. Informatics and data analytics to support exposome-based discovery for public health. Annual Review of Public Health 38:279–294. https://doi.org/10.1146/annurev-publhealth-082516-012737.
Marks, K. E., and D. A. Rao. 2022. T peripheral helper cells in autoimmune diseases. Immunological Reviews. https://doi.org/10.1111/imr.13069.
Márquez, A., M. Kerick, A. Zhernakova, J. Gutierrez-Achury, W.-M. Chen, S. Onengut-Gumuscu, I. González-Álvaro, L. Rodriguez-Rodriguez, R. Rios-Fernández, M. A. González-Gay, Coeliac Disease Immunochip Consortium, Rheumatoid Arthritis Consortium International for Immunochip (RACI), International Scleroderma Group, Type 1 Diabetes Genetics Consortium, M. D. Mayes, S. Raychaudhuri, S. S. Rich, C. Wijmenga, and J. Martín. 2018. Meta-analysis of immunochip data of four autoimmune diseases reveals novel single-disease and cross-phenotype associations. Genome Medicine 10(1):97. https://doi.org/10.1186/s13073-018-0604-8.
Martinsen, V., and P. Kursula. 2022. Multiple sclerosis and myelin basic protein: Insights into protein disorder and disease. Amino Acids 54(1):99–109. https://doi.org/10.1007/s00726-021-03111-7.
Martz, C. D., E. A. Hunter, M. R. Kramer, Y. Wang, K. Chung, M. Brown, C. Drenkard, S. S. Lim, and D. H. Chae. 2021. Pathways linking census tract typologies with subjective neighborhood disorder and depressive symptoms in the Black women’s experiences living with lupus (BeWELL) study. Health & Place 70:102587. https://doi.org/10.1016/j.healthplace.2021.102587.
Marx, A., Y. Yamada, K. Simon-Keller, B. Schalke, N. Willcox, P. Ströbel, and C.-A. Weis. 2021. Thymus and autoimmunity. Seminars in Immunopathology 43(1):45–64. https://doi.org/10.1007/s00281-021-00842-3.
Matzaraki, V., V. Kumar, C. Wijmenga, and A. Zhernakova. 2017. The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biology 18(1):76. https://doi.org/10.1186/s13059-017-1207-1.
Mazloom, S. E., D. Yan, J. Z. Hu, J. Ya, M. E. Husni, C. B. Warren, and A. P. Fernandez. 2020. TNF-α inhibitor-induced psoriasis: A decade of experience at the Cleveland clinic. Journal of the American Academy of Dermatology 83(6):1590–1598. https://doi.org/10.1016/j.jaad.2018.12.018.
Mazzone, R., C. Zwergel, M. Artico, S. Taurone, M. Ralli, A. Greco, and A. Mai. 2019. The emerging role of epigenetics in human autoimmune disorders. Clinical Epigenetics 11(1):34. https://doi.org/10.1186/s13148-019-0632-2.
McClain, M. T., L. D. Heinlen, G. J. Dennis, J. Roebuck, J. B. Harley, and J. A. James. 2005. Early events in lupus humoral autoimmunity suggest initiation through molecular mimicry. Nature Medicine 11(1):85–89. https://doi.org/10.1038/nm1167.
Mehri, F., E. Jenabi, S. Bashirian, F. G. Shahna, and S. Khazaei. 2020. The association between occupational exposure to silica and risk of developing rheumatoid arthritis: A meta-analysis. Safety and Health at Work 11(2):136–142. https://doi.org/10.1016/j.shaw.2020.02.001.
Mehta, P., R. Q. Cron, J. Hartwell, J. J. Manson, and R. S. Tattersall. 2020. Silencing the cytokine storm: The use of intravenous anakinra in haemophagocytic lymphohistiocytosis or macrophage activation syndrome. The Lancet Rheumatology 2(6):e358–e367. https://doi.org/10.1016/s2665-9913(20)30096-5.
Mentis, A. A., E. Dardiotis, N. Grigoriadis, E. Petinaki, and G. M. Hadjigeorgiou. 2017. Viruses and endogenous retroviruses in multiple sclerosis: From correlation to causation. Acta Neurologica Scandinavica 136(6):606–616. https://doi.org/10.1111/ane.12775.
Meyer, B., R. A. Chavez, J. E. Munro, R. C. Chiaroni-Clarke, J. D. Akikusa, R. C. Allen, J. M. Craig, A. L. Ponsonby, R. Saffery, and J. A. Ellis. 2015. DNA methylation at IL32 in juvenile idiopathic arthritis. Scientific Reports 5:11063. https://doi.org/10.1038/srep11063.
Miettinen, M. E., S. Niinisto, I. Erlund, D. Cuthbertson, A. M. Nucci, J. Honkanen, O. Vaarala, H. Hyoty, J. P. Krischer, M. Knip, S. M. Virtanen, and TRIGR Investigators. 2020. Serum 25-hydroxyvitamin D concentration in childhood and risk of islet autoimmunity and type 1 diabetes: The TRIGR nested case-control ancillary study. Diabetologia 63(4):780–787. https://doi.org/10.1007/s00125-019-05077-4.
Miller, F. W., L. Alfredsson, K. H. Costenbader, D. L. Kamen, L. M. Nelson, J. M. Norris, and A. J. De Roos. 2012. Epidemiology of environmental exposures and human autoimmune diseases: Findings from a national institute of environmental health sciences expert panel workshop. Journal of Autoimmunity 39(4):259–271. https://doi.org/10.1016/j.jaut.2012.05.002.
Mingueneau, M., S. Boudaoud, S. Haskett, T. L. Reynolds, G. Nocturne, E. Norton, X. Zhang, M. Constant, D. Park, W. Wang, T. Lazure, C. Le Pajolec, A. Ergun, and X. Mariette. 2016. Cytometry by time-of-flight immunophenotyping identifies a blood Sjögren’s signature correlating with disease activity and glandular inflammation. Journal of Allergy and Clinical Immunology 137(6):1809–1821 e1812. https://doi.org/10.1016/j.jaci.2016.01.024.
Mohammad Hosseini, A., J. Majidi, B. Baradaran, and M. Yousefi. 2015. Toll-like receptors in the pathogenesis of autoimmune diseases. Advanced Pharmaceutical Bulletin 5(Suppl 1):605–614. https://doi.org/10.15171/apb.2015.082.
Mori, K., and K. Yoshida. 2010. Viral infection in induction of Hashimoto’s thyroiditis: A key player or just a bystander? Current Opinion in Endocrinology, Diabetes, and Obesity 17(5):418–424. https://doi.org/10.1097/MED.0b013e32833cf518.
Morotti, A., I. Sollaku, S. Catalani, F. Franceschini, I. Cavazzana, M. Fredi, E. Sala, and G. De Palma. 2021. Systematic review and meta-analysis of epidemiological studies on the association of occupational exposure to free crystalline silica and systemic lupus erythematosus. Rheumatology (Oxford) 60(1):81–91. https://doi.org/10.1093/rheumatology/keaa444.
Morris, D. L., Y. Sheng, Y. Zhang, Y.-F. Wang, Z. Zhu, P. Tombleson, L. Chen, D. S. Cunninghame Graham, J. Bentham, A. L. Roberts, R. Chen, X. Zuo, T. Wang, L. Wen, C. Yang, L. Liu, L. Yang, F. Li, Y. Huang, X. Yin, S. Yang, L. Rönnblom, B. G. Fürnrohr, R. E. Voll, G. Schett, N. Costedoat-Chalumeau, P. M. Gaffney, Y. L. Lau, X. Zhang, W. Yang, Y. Cui, and T. J. Vyse. 2016. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nature Genetics 48(8):940–946. https://doi.org/10.1038/ng.3603.
Moulton, V. R. 2018. Sex hormones in acquired immunity and autoimmune disease. Frontiers in Immunology 9:2279. https://doi.org/10.3389/fimmu.2018.02279.
Munger, K. L., L. I. Levin, B. W. Hollis, N. S. Howard, and A. Ascherio. 2006. Serum 25-hy-droxyvitamin D levels and risk of multiple sclerosis. JAMA 296(23):2832–2838. https://doi.org/10.1001/jama.296.23.2832.
Munoz-Grajales, C., S. D. Prokopec, S. R. Johnson, Z. Touma, Z. Ahmad, D. Bonilla, L. Hiraki, A. Bookman, P. C. Boutros, A. Chruscinski, and J. Wither. 2021. Serological abnormalities that predict progression to systemic autoimmune rheumatic diseases in antinuclear antibody positive individuals. Rheumatology (Oxford). https://doi.org/10.1093/rheumatology/keab501.
Myers, J. M., D. Fairweather, S. A. Huber, and M. W. Cunningham. 2013. Autoimmune myocarditis, valvulitis, and cardiomyopathy. Current Protocols in Immunology Chapter 15:Unit 15.14.11–51. https://doi.org/10.1002/0471142735.im1514s101.
Myers, J. M., L. T. Cooper, D. C. Kem, S. Stavrakis, S. D. Kosanke, E. M. Shevach, D. Fairweather, J. A. Stoner, C. J. Cox, and M. W. Cunningham. 2016. Cardiac myosin-Th17 responses promote heart failure in human myocarditis. JCI Insight 1(9). https://doi.org/10.1172/jci.insight.85851.
Nelson, B. H. 2004. Il-2, regulatory t cells, and tolerance. Journal of Immunology 172(7):3983–3988. https://doi.org/10.4049/jimmunol.172.7.3983.
Ng, K., H. Stavropoulos, V. Anand, R. Veijola, J. Toppari, M. Maziarz, M. Lundgren, K. Waugh, B. I. Frohnert, F. Martin, W. Hagopian, and P. Achenbach. 2022. Islet autoantibody type-specific titer thresholds improve stratification of risk of progression to type 1 diabetes in children. Diabetes Care 45(1):160–168. https://doi.org/10.2337/dc21-0878.
Ng, S. C., W. Tang, R. W. Leong, M. Chen, Y. Ko, C. Studd, O. Niewiadomski, S. Bell, M. A. Kamm, H. J. de Silva, A. Kasturiratne, Y. U. Senanayake, C. J. Ooi, K.-L. Ling, D. Ong, K. L. Goh, I. Hilmi, Q. Ouyang, Y.-F. Wang, P. Hu, Z. Zhu, Z. Zeng, K. Wu, X. Wang, B. Xia, J. Li, P. Pisespongsa, S. Manatsathit, S. Aniwan, M. Simadibrata, M. Abdullah, S. W. Tsang, T. C. Wong, A. J. Hui, C. M. Chow, H. H. Yu, M. F. Li, K. K. Ng, J. Ching, J. C. Wu, F. K. Chan, and J. J. Sung, and on behalf of the Asia-Pacific Crohn’s and Colitis Colitis Epidemiology Study (ACCESS) Group. 2015. Environmental risk factors in inflammatory bowel disease: A population-based case-control study in Asia-Pacific. Gut 64(7):1063–1071. https://doi.org/10.1136/gutjnl-2014-307410.
NHGRI (National Human Genome Research Institute). n.d. Immune system disease: Genome-wide association study (GWAS), edited by National Human Genome Research Institute: EMBL-EBI.
NHGRI. 2020. Polygenic risk scores. https://www.genome.gov/Health/Genomics-and-Medicine/Polygenic-risk-scores (accessed February 16, 2022).
NIH (National Institutes of Health). 2021. Clinicaltrials.Gov is a database of privately and publicly funded clinical studies conducted around the world. https://clinicaltrials.gov (accessed May 8, 2021).
Nistico, L., D. Iafusco, A. Galderisi, C. Fagnani, R. Cotichini, V. Toccaceli, M. A. Stazi, E. Study Group on Diabetes of the Italian Society of Pediatric, and Diabetology. 2012. Emerging effects of early environmental factors over genetic background for type 1 diabetes susceptibility: Evidence from a nationwide Italian twin study. Journal of Clinical Endocrinology & Metabolism 97(8):E1483–1491. https://doi.org/10.1210/jc.2011-3457.
Noonan, C. W., J. C. Pfau, T. C. Larson, and M. R. Spence. 2006. Nested case-control study of autoimmune disease in an asbestos-exposed population. Environmental Health Perspectives 114(8):1243–1247. https://doi.org/10.1289/ehp.9203.
Norris, J. M., R. K. Johnson, and L. C. Stene. 2020. Type 1 diabetes-early life origins and changing epidemiology. The Lancet Diabetes & Endocrinology 8(3):226–238. https://doi.org/10.1016/S2213-8587(19)30412-7.
Notkins, A. L. 2004. Type 1 diabetes as a model for autoantibodies as predictors of autoimmune diseases. Autoimmunity Reviews 3(Suppl 1):S7–9.
Nowak, K., E. Jabłońska, and W. Ratajczak-Wrona. 2019. Immunomodulatory effects of synthetic endocrine disrupting chemicals on the development and functions of human immune cells. Environment International 125:350–364. https://doi.org/10.1016/j.envint.2019.01.078.
O’Shea, J. 2021. NIAMS IRP 101. Paper read at NASEM Committee for the Assessment of NIH Research on Autoimmune Diseases: Meeting #3, March 4, 2021, Webinar.
Okada, Y., D. Wu, G. Trynka, T. Raj, C. Terao, K. Ikari, Y. Kochi, K. Ohmura, A. Suzuki, S. Yoshida, R. R. Graham, A. Manoharan, W. Ortmann, T. Bhangale, J. C. Denny, R. J. Carroll, A. E. Eyler, J. D. Greenberg, J. M. Kremer, D. A. Pappas, L. Jiang, J. Yin, L. Ye, D.-F. Su, J. Yang, G. Xie, E. Keystone, H.-J. Westra, T. Esko, A. Metspalu, X. Zhou, N. Gupta, D. Mirel, E. A. Stahl, D. Diogo, J. Cui, K. Liao, M. H. Guo, K. Myouzen, T. Kawaguchi, M. J. Coenen, P. L. van Riel, M. A. van de Laar, H.-J. Guchelaar, T. W. Huizinga, P. Dieudé, X. Mariette, S. L. Bridges, Jr., A. Zhernakova, R. E. Toes, P. P. Tak, C. Miceli-Richard, S.-Y. Bang, H.-S. Lee, J. Martin, M. A. Gonzalez-Gay, L. Rodriguez-Rodriguez, S. Rantapää-Dahlqvist, L. Ärlestig, H. K. Choi, Y. Kamatani, P. Galan, M. Lathrop, the RACI consortium, the GARNET consortium, S. Eyre, J. Bowes, A. Barton, N. de Vries, L. W. Moreland, L. A. Criswell, E. W. Karlson, A. Taniguchi, R. Yamada, M. Kubo, J. S. Liu, S.-C. Bae, J. Worthington, L. Padyukov, L. Klareskog, P. K. Gregersen, S. Raychaudhuri, B. E. Stranger, P. L. De Jager, L. Franke, P. M. Visscher, M. A. Brown, H. Yamanaka, T. Mimori, A. Takahashi, H. Xu, T. W. Behrens, K. A. Siminovitch, S. Momohara, F. Matsuda, K. Yamamoto, and R. M. Plenge. 2014. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506(7488):376–381. https://doi.org/10.1038/nature12873.
Onengut-Gumuscu, S., W.-M. Chen, O. Burren, N. J. Cooper, A. R. Quinlan, J. C. Mychaleckyj, E. Farber, J. K. Bonnie, M. Szpak, E. Schofield, P. Achuthan, H. Guo, M. D. Fortune, H. Stevens, N. M. Walker, L. D. Ward, A. Kundaje, M. Kellis, M. J. Daly, J. C. Barrett, J. D. Cooper, P. Deloukas, Type 1 Diabetes Genetics Consortium, J. A. Todd, C. Wallace, P. Concannon, and S. S. Rich. 2015. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nature Genetics 47(4):381–386. https://doi.org/10.1038/ng.3245.
Palla, G., and E. Ferrero. 2020. Latent factor modeling of scrna-seq data uncovers dysregulated pathways in autoimmune disease patients. iScience 23(9):101451. https://doi.org/10.1016/j.isci.2020.101451.
Pappalardo, F., G. Russo, F. M. Tshinanu, and M. Viceconti. 2019. In silico clinical trials: Concepts and early adoptions. Briefings in Bioinformatics 20(5):1699–1708. https://doi.org/10.1093/bib/bby043.
Parisis, D., C. Bernier, F. Chasset, and L. Arnaud. 2019. Impact of tobacco smoking upon disease risk, activity and therapeutic response in systemic lupus erythematosus: A systematic review and meta-analysis. Autoimmunity Reviews 18(11):102393.
Park, C. Y., and S. N. Han. 2021. The role of vitamin d in adipose tissue biology: Adipocyte differentiation, energy metabolism, and inflammation. Journal of Lipid and Atherosclerosis 10(2):130–144. https://doi.org/10.12997/jla.2021.10.2.130.
Park, D., J. Lee, J. J. Chung, Y. Jung, and S. H. Kim. 2020. Integrating organs-on-chips: Multiplexing, scaling, vascularization, and innervation. Trends in Biotechnology 38(1):99–112. https://doi.org/10.1016/j.tibtech.2019.06.006.
Parks, C. G., K. Conrad, and G. S. Cooper. 1999. Occupational exposure to crystalline silica and autoimmune disease. Environmental Health Perspectives 107(Suppl 5):793–802. https://doi.org/10.1289/ehp.99107s5793.
Parks, C. G., F. W. Miller, K. M. Pollard, C. Selmi, D. Germolec, K. Joyce, N. R. Rose, and M. C. Humble. 2014. Expert panel workshop consensus statement on the role of the environment in the development of autoimmune disease. International Journal of Molecular Sciences 15(8):14269–14297. https://doi.org/10.3390/ijms150814269.
Pattison, D. J., D. P. M. Symmons, M. Lunt, A. Welch, R. Luben, S. A. Bingham, K.-T. Khaw, N. E. Day, and A. J. Silman. 2004. Dietary risk factors for the development of inflammatory polyarthritis: Evidence for a role of high level of red meat consumption. Arthritis and Rheumatism 50(12):3804–3812. https://doi.org/10.1002/art.20731.
Pavlovic, K., M. Tristán-Manzano, N. Maldonado-Pérez, M. Cortijo-Gutierrez, S. Sánchez-Hernández, P. Justicia-Lirio, M. D. Carmona, C. Herrera, F. Martin, and K. Benabdellah. 2020. Using gene editing approaches to fine-tune the immune system. Frontiers in Immunology 11:570672. https://doi.org/10.3389/fimmu.2020.570672.
Persson, L., S. Longhi, J. Enarsson, O. Andersen, S. Haghigi, S. Nilsson, M. Lagging, M. Johansson, and T. Bergstrom. 2014. Elevated antibody reactivity to measles virus ncore protein among patients with multiple sclerosis and their healthy siblings with intrathe-cal oligoclonal immunoglobulin g production. Journal of Clinical Virology 61(1):107–112. https://doi.org/10.1016/j.jcv.2014.06.011.
Pfau, J. C., C. Barbour, B. Black, K. M. Serve, and M. J. Fritzler. 2018. Analysis of autoantibody profiles in two asbestiform fiber exposure cohorts. Journal of Toxicology and Environmental Health. Part A 81(19):1015–1027. https://doi.org/10.1080/15287394.2018.1512432.
Piccinni, M. P., R. Raghupathy, S. Saito, and J. Szekeres-Bartho. 2021. Cytokines, hormones and cellular regulatory mechanisms favoring successful reproduction. Frontiers in Immunology 12(2944):717808. https://doi.org/10.3389/fimmu.2021.717808.
Piovani, D., C. Pansieri, S. R. R. Kotha, A. C. Piazza, C.-L. Comberg, L. Peyrin-Biroulet, S. Danese, and S. Bonovas. 2021. Ethnic differences in the smoking-related risk of inflammatory bowel disease: A systematic review and meta-analysis. Journal of Crohn’s & Colitis 15(10):1658–1678. https://doi.org/10.1093/ecco-jcc/jjab047.
Polini, A., L. L. Del Mercato, A. Barra, Y. S. Zhang, F. Calabi, and G. Gigli. 2019. Towards the development of human immune-system-on-a-chip platforms. Drug Discovery Today 24(2):517–525. https://doi.org/10.1016/j.drudis.2018.10.003.
Popescu, M., T. B. Feldman, and T. Chitnis. 2021. Interplay between endocrine disruptors and immunity: Implications for diseases of autoreactive etiology. Frontiers in Pharmacology 12:626107. https://doi.org/10.3389/fphar.2021.626107.
Primavera, M., C. Giannini, and F. Chiarelli. 2020. Prediction and prevention of type 1 diabetes. Frontiers in Endocrinology 11:248. https://doi.org/10.3389/fendo.2020.00248.
Psarras, A., P. Emery, and E. M. Vital. 2017. Type I interferon-mediated autoimmune diseases: Pathogenesis, diagnosis and targeted therapy. Rheumatology (Oxford) 56(10):1662–1675. https://doi.org/10.1093/rheumatology/kew431.
Qiu, F., R. Tang, X. Zuo, X. Shi, Y. Wei, X. Zheng, Y. Dai, Y. Gong, L. Wang, P. Xu, X. Zhu, J. Wu, C. Han, Y. Gao, K. Zhang, Y. Jiang, J. Zhou, Y. Shao, Z. Hu, Y. Tian, H. Zhang, N. Dai, L. Liu, X. Wu, W. Zhao, X. Zhang, Z. Zang, J. Nie, W. Sun, Y. Zhao, Y. Mao, P. Jiang, H. Ji, Q. Dong, J. Li, Z. Li, X. Bai, L. Li, M. Lin, M. Dong, J. Li, P. Zhu, C. Wang, Y. Zhang, P. Jiang, Y. Wang, R. Jawed, J. Xu, Y. Zhang, Q. Wang, Y. Yang, F. Yang, M. Lian, X. Jiang, X. Xiao, Y. Li, J. Fang, D. Qiu, Z. Zhu, H. Qiu, J. Zhang, W. Tian, S. Chen, L. Jiang, B. Ji, P. Li, G. Chen, T. Wu, Y. Sun, J. Yu, H. Tang, M. He, M. Xia, H. Pei, L. Huang, Z. Qing, J. Wu, Q. Huang, J. Han, W. Xie, Z. Sun, J. Guo, G. He, M. Eric Gershwin, Z. Lian, X. Liu, M. F. Seldin, X. Liu, W. Chen, and X. Ma. 2017. A genome-wide association study identifies six novel risk loci for primary biliary cholangitis. Nature Communications 8:14828. https://doi.org/10.1038/ncomms14828.
Racine, A., F. Carbonnel, S. S. M. Chan, A. R. Hart, H. B. Bueno-de-Mesquita, B. Oldenburg, F. D. M. van Schaik, A. Tjønneland, A. Olsen, C. C. Dahm, T. Key, R. Luben, K.-T. Khaw, E. Riboli, O. Grip, S. Lindgren, G. Hallmans, P. Karling, F. Clavel-Chapelon, M. M. Bergman, H. Boeing, R. Kaaks, V. A. Katzke, D. Palli, G. Masala, P. Jantchou, and M.-C. Boutron-Ruault. 2016. Dietary patterns and risk of inflammatory bowel disease in Europe: Results from the EPIC study. Inflammatory Bowel Diseases 22(2):345–354. https://doi.org/10.1097/MIB.0000000000000638.
Radstake, T. R. D. J., O. Gorlova, B. Rueda, J.-E. Martin, B. Z. Alizadeh, R. Palomino-Morales, M. J. Coenen, M. C. Vonk, A. E. Voskuyl, A. J. Schuerwegh, J. C. Broen, P. L. C. M. van Riel, R. van ‘t Slot, A. Italiaander, R. A. Ophoff, G. Riemekasten, N. Hunzelmann, C. P. Simeon, N. Ortego-Centeno, M. A. González-Gay, M. F. González-Escribano, Spanish Scleroderma Group, P. Airo, J. van Laar, A. Herrick, J. Worthington, R. Hesselstrand, V. Smith, F. de Keyser, F. Houssiau, M. M. Chee, R. Madhok, P. Shiels, R. Westhovens, A. Kreuter, H. Kiener, E. de Baere, T. Witte, L. Padykov, L. Klareskog, L. Beretta, R. Scorza, B. A. Lie, A. M. Hoffmann-Vold, P. Carreira, J. Varga, M. Hinchcliff, P. K. Gregersen, A. T. Lee, J. Ying, Y. Han, S. F. Weng, C. I. Amos, F. M. Wigley, L. Hummers, J. L. Nelson, S. K. Agarwal, S. Assassi, P. Gourh, F. K. Tan, B. P. Koeleman, F. C. Arnett, J. Martin, and M. D. Mayes. 2010. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nature Genetics 42(5):426–429.
Ramaswamy, M., R. Tummala, K. Streicher, A. Nogueira da Costa, and P. Z. Brohawn. 2021. The pathogenesis, molecular mechanisms, and therapeutic potential of the interferon pathway in systemic lupus erythematosus and other autoimmune diseases. International Journal of Molecular Sciences 22(20). https://doi.org/10.3390/ijms222011286.
Rapisarda, V., C. Loreto, S. Castorina, G. Romano, S. F. Garozzo, A. Musumeci, M. Migliore, R. Avola, D. Cina, C. Pomara, and C. Ledda. 2018. Occupational exposure to fluoroedenite and prevalence of anti-nuclear autoantibodies. Future Oncology (London, England) 14(6s):59–62. https://doi.org/10.2217/fon-2017-0389.
Regitz-Zagrosek, V., E. Becher, S. Mahmoodzadeh, and C. Schubert. 2008. Sex steroid hormones. In Cardiovascular hormone systems: From molecular mechanisms to novel therapeutics, edited by M. Bader. Weinheim, Germany: John Wiley & Sons. Pp. 39–64.
Reichlin, M., J. B. Harley, and M. D. Lockshin. 1992. Serologic studies of monozygotic twins with systemic lupus erythematosus. Arthritis and Rheumatism 35(4):457–464. https://doi.org/10.1002/art.1780350416.
Reid, A., P. Franklin, N. de Klerk, J. Creaney, F. Brims, B. Musk, and J. Pfau. 2018. Autoimmune antibodies and asbestos exposure: Evidence from Wittenoom, Western Australia. American Journal of Industrial Medicine 61(7):615–620. https://doi.org/10.1002/ajim.22863.
Reid, S., A. Alexsson, M. Frodlund, D. Morris, J. K. Sandling, K. Bolin, E. Svenungsson, A. Jonsen, C. Bengtsson, I. Gunnarsson, V. Illescas Rodriguez, A. Bengtsson, S. Arve, S. Rantapaa-Dahlqvist, M. L. Eloranta, A. C. Syvanen, C. Sjowall, T. J. Vyse, L. Ronnblom, and D. Leonard. 2020. High genetic risk score is associated with early disease onset, damage accrual and decreased survival in systemic lupus erythematosus. Annals of the Rheumatic Diseases 79(3):363–369. https://doi.org/10.1136/annrheumdis-2019-216227.
Richard-Miceli, C., and L. A. Criswell. 2012. Emerging patterns of genetic overlap across autoimmune disorders. Genome Medicine 4(1):6. https://doi.org/10.1186/gm305.
Ristevski, B., and M. Chen. 2018. Big data analytics in medicine and healthcare. Journal of Integrative Bioinformatics 15(3). https://doi.org/10.1515/jib-2017-0030.
Robles-Matos, N., T. Artis, R. A. Simmons, and M. S. Bartolomei. 2021. Environmental exposure to endocrine disrupting chemicals influences genomic imprinting, growth, and metabolism. Genes (Basel) 12(8). https://doi.org/10.3390/genes12081153.
Rodriguez-Calvo, T. 2019. Enterovirus infection and type 1 diabetes: Unraveling the crime scene. Clinical & Experimental Immunology 195(1):15–24. https://doi.org/10.1111/cei.13223.
Root-Bernstein, R., and D. Fairweather. 2015. Unresolved issues in theories of autoimmune disease using myocarditis as a framework. Journal of Theoretical Biology 375:101–123. https://doi.org/10.1016/j.jtbi.2014.11.022.
Rubio-Rivas, M., R. Moreno, and X. Corbella. 2017. Occupational and environmental scleroderma. Systematic review and meta-analysis. Clinical Rheumatology 36(3):569–582. https://doi.org/10.1007/s10067-016-3533-1.
Ruchakorn, N., P. Ngamjanyaporn, T. Suangtamai, T. Kafaksom, C. Polpanumas, V. Petpisit, T. Pisitkun, and P. Pisitkun. 2019. Performance of cytokine models in predicting SLE activity. Arthritis Research & Therapy 21(1):287. https://doi.org/10.1186/s13075-019-2029-1.
Saevarsdottir, S., T. A. Olafsdottir, E. V. Ivarsdottir, G. H. Halldorsson, K. Gunnarsdottir, A. Sigurdsson, A. Johannesson, J. K. Sigurdsson, T. Juliusdottir, S. H. Lund, A. O. Arnthorsson, E. L. Styrmisdottir, J. Gudmundsson, G. M. Grondal, K. Steinsson, L. Alfredsson, J. Askling, R. Benediktsson, R. Bjarnason, A. J. Geirsson, B. Gudbjorns-son, H. Gudjonsson, H. Hjaltason, A. B. Hreidarsson, L. Klareskog, I. Kockum, H. Kristjansdottir, T. J. Love, B. R. Ludviksson, T. Olsson, P. T. Onundarson, K. B. Orvar, L. Padyukov, B. Sigurgeirsson, V. Tragante, K. Bjarnadottir, T. Rafnar, G. Masson, P. Sulem, D. F. Gudbjartsson, P. Melsted, G. Thorleifsson, G. L. Norddahl, U. Thorsteinsdottir, I. Jonsdottir, and K. Stefansson. 2020. FLT3 stop mutation increases FLT3 ligand level and risk of autoimmune thyroid disease. Nature 584(7822):619–623. https://doi.org/10.1038/s41586-020-2436-0.
Sandling, J. K., P. Pucholt, L. Hultin Rosenberg, F. H. G. Farias, S. V. Kozyrev, M. L. Eloranta, A. Alexsson, M. Bianchi, L. Padyukov, C. Bengtsson, R. Jonsson, R. Omdal, B. A. Lie, L. Massarenti, R. Steffensen, M. A. Jakobsen, S. T. Lillevang, on behalf of the ImmunoArray Development Consortium and DISSECT consortium, K. Lerang, O. Molberg, A. Voss, A. Troldborg, S. Jacobsen, A. C. Syvanen, A. Jonsen, I. Gunnarsson, E. Svenungsson, S. Rantapaa-Dahlqvist, A. A. Bengtsson, C. Sjowall, D. Leonard, K. Lindblad-Toh, and L. Ronnblom. 2021. Molecular pathways in patients with systemic lupus erythematosus revealed by gene-centred DNA sequencing. Annals of the Rheumatic Diseases 80(1):109–117. https://doi.org/10.1136/annrheumdis-2020-218636.
Sawcer, S., G. Hellenthal, M. Pirinen, C. C. A. Spencer, N. A. Patsopoulos, L. Moutsianas, A. Dilthey, Z. Su, C. Freeman, S. E. Hunt, S. Edkins, E. Gray, D. R. Booth, S. C. Potter, A. Goris, G. Band, A. B. Oturai, A. Strange, J. Saarela, C. Bellenguez, B. Fontaine, M. Gillman, B. Hemmer, R. Gwilliam, F. Zipp, A. Jayakumar, R. Martin, S. Leslie, S. Hawkins, E. Giannoulatou, S. D’Alfonso, H. Blackburn, F. Martinelli Boneschi, J. Liddle, H. F. Harbo, M. L. Perez, A. Spurkland, M. J. Waller, M. P. Mycko, M. Ricketts, M. Comabella, N. Hammond, I. Kockum, O. T. McCann, M. Ban, P. Whittaker, A. Kemppinen, P. Weston, C. Hawkins, S. Widaa, J. Zajicek, S. Dronov, N. Robertson, S. J. Bumpstead, L. F. Barcellos, R. Ravindrarajah, R. Abraham, L. Alfredsson, K. Ardlie, C. Aubin, A. Baker, K. Baker, S. E. Baranzini, L. Bergamaschi, R. Bergamaschi, A. Bernstein, A. Berthele, M. Boggild, J. P. Bradfield, D. Brassat, S. A. Broadley, D. Buck, H. Butzkueven, R. Capra, W. M. Carroll, P. Cavalla, E. G. Celius, S. Cepok, R. Chiavacci, F. Clerget-Darpoux, K. Clysters, G. Comi, M. Cossburn, I. Cournu-Rebeix, M. B. Cox, W. Cozen, B. A. Cree, A. H. Cross, D. Cusi, M. J. Daly, E. Davis, P. I. de Bakker, M. Debouverie, B. D’Hooghe M, K. Dixon, R. Dobosi, B. Dubois, D. Ellinghaus, I. Elovaara, F. Esposito, C. Fontenille, S. Foote, A. Franke, D. Galimberti, A. Ghezzi, J. Glessner, R. Gomez, O. Gout, C. Graham, S. F. Grant, F. R. Guerini, H. Hakonarson, P. Hall, A. Hamsten, H. P. Hartung, R. N. Heard, S. Heath, J. Hobart, M. Hoshi, C. Infante-Duarte, G. Ingram, W. Ingram, T. Islam, M. Jagodic, M. Kabesch, A. G. Kermode, T. J. Kilpatrick, C. Kim, N. Klopp, K. Koivisto, M. Larsson, M. Lathrop, J. S. Lechner-Scott, M. A. Leone, V. Leppä, U. Liljedahl, I. L. Bomfim, R. R. Lincoln, J. Link, J. Liu, A. R. Lorentzen, S. Lupoli, F. Macciardi, T. Mack, M. Marriott, V. Martinelli, D. Mason, J. L. McCauley, F. Mentch, I. L. Mero, T. Mihalova, X. Montalban, J. Mottershead, K. M. Myhr, P. Naldi, W. Ollier, A. Page, A. Palotie, J. Pelletier, L. Piccio, T. Pickersgill, F. Piehl, S. Pobywajlo, H. L. Quach, P. P. Ramsay, M. Reunanen, R. Reynolds, J. D. Rioux, M. Rodegher, S. Roesner, J. P. Rubio, I. M. Rückert, M. Salvetti, E. Salvi, A. Santaniello, C. A. Schaefer, S. Schreiber, C. Schulze, R. J. Scott, F. Sellebjerg, K. W. Selmaj, D. Sexton, L. Shen, B. Simms-Acuna, S. Skidmore, P. M. Sleiman, C. Smestad, P. S. Sørensen, H. B. Søndergaard, J. Stankovich, R. C. Strange, A. M. Sulonen, E. Sundqvist, A. C. Syvänen, F. Taddeo, B. Taylor, J. M. Blackwell, P. Tienari, E. Bramon, A. Tourbah, M. A. Brown, E. Tronczynska, J. P. Casas, N. Tubridy, A. Corvin, J. Vickery, J. Jankowski, P. Villoslada, H. S. Markus, K. Wang, C. G. Mathew, J. Wason, C. N. Palmer, H. E. Wichmann, R. Plomin, E. Willoughby, A. Rautanen, J. Winkelmann, M. Wittig, R. C. Trembath, J. Yaouanq, A. C. Viswanathan, H. Zhang, N. W. Wood, R. Zuvich, P. Deloukas, C. Langford, A. Duncanson, J. R. Oksenberg, M. A. Pericak-Vance, J. L. Haines, T. Olsson, J. Hillert, A. J. Ivinson, P. L. De Jager, L. Peltonen, G. J. Stewart, D. A. Hafler, S. L. Hauser, G. McVean, P. Donnelly, and A. Compston. 2011. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476(7359):214–219. https://doi.org/10.1038/nature10251.
Schlegel, A., C. Wang, B. S. Katzenellenbogen, R. G. Pestell, and M. P. Lisanti. 1999. Caveolin-1 potentiates estrogen receptor alpha (ERalpha) signaling. Caveolin-1 drives ligand-independent nuclear translocation and activation of eralpha. Journal of Biological Chemistry 274(47):33551–33556. https://doi.org/10.1074/jbc.274.47.33551.
Shamim, E. A., and F. W. Miller. 2000. Familial autoimmunity and the idiopathic inflammatory myopathies. Current Rheumatology Reports 2(3):201–211. https://doi.org/10.1007/s11926-000-0080-0.
Sharp, S. A., M. N. Weedon, W. A. Hagopian, and R. A. Oram. 2018. Clinical and research uses of genetic risk scores in type 1 diabetes. Current Opinion in Genetics & Development 50:96–102. https://doi.org/10.1016/j.gde.2018.03.009.
Simmons, S. B., S. Schippling, G. Giovannoni, and D. Ontaneda. 2021. Predicting disability worsening in relapsing and progressive multiple sclerosis. Current Opinion in Neurology 34(3):312–321. https://doi.org/10.1097/WCO.0000000000000928.
Sogkas, G., F. Atschekzei, I. R. Adriawan, N. Dubrowinskaja, T. Witte, and R. E. Schmidt. 2021. Cellular and molecular mechanisms breaking immune tolerance in inborn errors of immunity. Cellular & Molecular Immunology 18(5):1122–1140. https://doi.org/10.1038/s41423-020-00626-z.
Somers, E. C., S. L. Thomas, L. Smeeth, and A. J. Hall. 2009. Are individuals with an autoimmune disease at higher risk of a second autoimmune disorder? American Journal of Epidemiology 169(6):749–755. https://doi.org/10.1093/aje/kwn408.
Stafford, I. S., M. Kellermann, E. Mossotto, R. M. Beattie, B. D. MacArthur, and S. Ennis. 2020. A systematic review of the applications of artificial intelligence and machine learning in autoimmune diseases. NPJ Digital Medicine 3:30. https://doi.org/10.1038/s41746-020-0229-3.
Stahl, E. A., S. Raychaudhuri, E. F. Remmers, G. Xie, S. Eyre, B. P. Thomson, Y. Li, F. A. S. Kurreeman, A. Zhernakova, A. Hinks, C. Guiducci, R. Chen, L. Alfredsson, C. I. Amos, K. G. Ardlie, BIRAC Consortium, A. Barton, J. Bowes, E. Brouwer, N. P. Burtt, J. J. Catanese, J. Coblyn, M. J. H. Coenen, K. H. Costenbader, L. A. Criswell, J. B. A. Crusius, J. Cui, P. I. W. de Bakker, P. L. De Jager, B. Ding, P. Emery, E. Flynn, P. Harrison, L. J. Hocking, T. W. Huizinga, D. L. Kastner, X. Ke, A. T. Lee, X. Liu, P. Martin, A. W. Morgan, L. Padyukov, M. D. Posthumus, T. R. Radstake, D. M. Reid, M. Seielstad, M. F. Seldin, N. A. Shadick, S. Steer, P. P. Tak, W. Thomson, A. H. van der Helm-van Mil, I. E. van der Horst-Bruinsma, C. E. van der Schoot, P. L. van Riel, M. E. Weinblatt, A. G. Wilson, G. J. Wolbink, B. P. Wordsworth, Y. Consortium, C. Wijmenga, E. W. Karlson, R. E. Toes, N. de Vries, A. B. Begovich, J. Worthington, K. A. Siminovitch, P. K. Gregersen, L. Klareskog, and R. M. Plenge. 2010. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nature Genetics 42(6):508–514. https://doi.org/10.1038/ng.582.
Surace, A. E. A., and C. M. Hedrich. 2019. The role of epigenetics in autoimmune/inflammatory disease. Frontiers in Immunology 10:1525. https://doi.org/10.3389/fimmu.2019.01525.
Svendsen, A. J., K. O. Kyvik, G. Houen, P. Junker, K. Christensen, L. Christiansen, C. Nielsen, A. Skytthe, and J. V. Hjelmborg. 2013. On the origin of rheumatoid arthritis: The impact of environment and genes—a population based twin study. PloS One 8(2):e57304. https://doi.org/10.1371/journal.pone.0057304.
Tam, P. E. 2006. Coxsackievirus myocarditis: Interplay between virus and host in the pathogenesis of heart disease. Viral Immunology 19(2):133–146. https://doi.org/10.1089/vim.2006.19.133.
Tanner, T., and D. M. Wahezi. 2020. Hyperinflammation and the utility of immunomodulatory medications in children with COVID-19. Paediatric Respiratory Reviews 35:81–87. https://doi.org/10.1016/j.prrv.2020.07.003.
Theander, E., R. Jonsson, B. Sjöström, K. Brokstad, P. Olsson, and G. Henriksson. 2015. Prediction of Sjögren’s syndrome years before diagnosis and identification of patients with early onset and severe disease course by autoantibody profiling. Arthritis & Rheumatology 67(9):2427–2436. https://doi.org/10.1002/art.39214.
Tintut, Y., and L. L. Demer. 2021. Potential impact of the steroid hormone, vitamin D, on the vasculature. American Heart Journal 239:147–153. https://doi.org/10.1016/j.ahj.2021.05.012.
Todd, J. A., N. M. Walker, J. D. Cooper, D. J. Smyth, K. Downes, V. Plagnol, R. Bailey, S. Nejentsev, S. F. Field, F. Payne, C. E. Lowe, J. S. Szeszko, J. P. Hafler, L. Zeitels, J. H. M. Yang, A. Vella, S. Nutland, H. E. Stevens, H. Schuilenburg, G. Coleman, M. Maisuria, W. Meadows, L. J. Smink, B. Healy, O. S. Burren, A. A. C. Lam, N. R. Ovington, J. Allen, E. Adlem, H.-T. Leung, C. Wallace, J. M. Howson, C. Guja, C. Ionescu-Tîrgovişte, Genetics of Type 1 Diabetes in Finland, M. J. Simmonds, J. M. Heward, S. C. Gough, D. B. Dunger, the Wellcome Trust Case Control Consortium, L. S. Wicker, and D. G. Clayton. 2007. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nature Genetics 39(7):857–864. https://doi.org/10.1038/ng2068.
Troncone, E., I. Marafini, G. Del Vecchio Blanco, A. Di Grazia, and G. Monteleone. 2020. Novel therapeutic options for people with ulcerative colitis: An update on recent developments with Janus kinase (JAK) inhibitors. Clinical and Experimental Gastroenterology 13:131–139. https://doi.org/10.2147/CEG.S208020.
Tsoi, L. C., S. L. Spain, J. Knight, E. Ellinghaus, P. E. Stuart, F. Capon, J. Ding, Y. Li, T. Tejasvi, J. E. Gudjonsson, H. M. Kang, M. H. Allen, R. McManus, G. Novelli, L. Samuelsson, J. Schalkwijk, M. Ståhle, A. D. Burden, C. H. Smith, M. J. Cork, X. Estivill, A. M. Bowcock, G. G. Krueger, W. Weger, J. Worthington, R. Tazi-Ahnini, F. O. Nestle, A. Hayday, P. Hoffmann, J. Winkelmann, C. Wijmenga, C. Langford, S. Edkins, R. Andrews, H. Blackburn, A. Strange, G. Band, R. D. Pearson, D. Vukcevic, C. C. Spencer, P. Deloukas, U. Mrowietz, S. Schreiber, S. Weidinger, S. Koks, K. Kingo, T. Esko, A. Metspalu, H. W. Lim, J. J. Voorhees, M. Weichenthal, H. E. Wichmann, V. Chandran, C. F. Rosen, P. Rahman, D. D. Gladman, C. E. Griffiths, A. Reis, J. Kere, Collaborative Association Study of Psoriasis (CASP), Genetic Analysis of Psoriasis Consortium, Psoriasis Association Genetics Extension, Wellcome Trust Case Control Consortium 2, R. P. Nair, A. Franke, J. N. Barker, G. R. Abecasis, J. T. Elder, and R. C. Trembath. 2012. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nature Genetics 44(12):1341–1348. https://doi.org/10.1038/ng.2467.
Tsoi, L. C., P. E. Stuart, C. Tian, J. E. Gudjonsson, S. Das, M. Zawistowski, E. Ellinghaus, J. N. Barker, V. Chandran, N. Dand, K. C. Duffin, C. Enerbäck, T. Esko, A. Franke, D. D. Gladman, P. Hoffmann, K. Kingo, S. Kõks, G. G. Krueger, H. W. Lim, A. Metspalu, U. Mrowietz, S. Mucha, P. Rahman, A. Reis, T. Tejasvi, R. Trembath, J. J. Voorhees, S. Weidinger, M. Weichenthal, X. Wen, N. Eriksson, H. M. Kang, D. A. Hinds, R. P. Nair, G. R. Abecasis, and J. T. Elder. 2017. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nature Communications 8:15382. https://doi.org/10.1038/ncomms15382.
Tucker, W. G., and R. Andrew Paskauskas. 2008. The msmv hypothesis: Measles virus and multiple sclerosis, etiology and treatment. Medical Hypotheses 71(5):682–689. https://doi.org/10.1016/j.mehy.2008.06.029.
Ulff-Møller, C. J., A. J. Svendsen, L. N. Viemose, and S. Jacobsen. 2018. Concordance of autoimmune disease in a nationwide Danish systemic lupus erythematosus twin cohort. Seminars in Arthritis and Rheumatism 47(4):538–544. https://doi.org/10.1016/j.semarthrit.2017.06.007.
Umiker, B. R., S. Andersson, L. Fernandez, P. Korgaokar, A. Larbi, M. Pilichowska, C. C. Weinkauf, H. H. Wortis, J. F. Kearney, and T. Imanishi-Kari. 2014. Dosage of x-linked toll-like receptor 8 determines gender differences in the development of systemic lupus erythematosus. European Journal of Immunology 44(5):1503–1516. https://doi.org/10.1002/eji.201344283.
Vallerand, I. A., R. T. Lewinson, A. D. Frolkis, M. W. Lowerison, G. G. Kaplan, M. G. Swain, A. G. M. Bulloch, S. B. Patten, and C. Barnabe. 2018. Depression as a risk factor for the development of rheumatoid arthritis: A population-based cohort study. RMD Open 4(2):e000670. https://doi.org/10.1136/rmdopen-2018-000670.
Vallerand, I. A., S. B. Patten, and C. Barnabe. 2019. Depression and the risk of rheumatoid arthritis. Current Opinion in Rheumatology 31(3):279–284. https://doi.org/10.1097/bor.0000000000000597.
van Pelt, E. D., J. Y. Mescheriakova, N. Makhani, I. A. Ketelslegers, R. F. Neuteboom, S. Kundu, L. Broer, C. Janssens, C. E. Catsman-Berrevoets, C. M. van Duijn, B. Banwell, A. Bar-Or, and R. Q. Hintzen. 2013. Risk genes associated with pediatric-onset MS but not with monophasic acquired CNS demyelination. Neurology 81(23):1996–2001. https://doi.org/10.1212/01.wnl.0000436934.40034eb.
Vestergaard, P. 2002. Smoking and thyroid disorders—a meta-analysis. European Journal of Endocrinology 146(2):153–161. https://doi.org/10.1530/eje.0.1460153.
Wang, K., R. Baldassano, H. Zhang, H. Q. Qu, M. Imielinski, S. Kugathasan, V. Annese, M. Dubinsky, J. I. Rotter, R. K. Russell, J. P. Bradfield, P. M. Sleiman, J. T. Glessner, T. Walters, C. Hou, C. Kim, E. C. Frackelton, M. Garris, J. Doran, C. Romano, C. Catassi, J. Van Limbergen, S. L. Guthery, L. Denson, D. Piccoli, M. S. Silverberg, C. A. Stanley, D. Monos, D. C. Wilson, A. Griffiths, S. F. Grant, J. Satsangi, C. Polychronakos, and H. Hakonarson. 2010. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Human Molecular Genetics 19(10):2059–2067. https://doi.org/10.1093/hmg/ddq078.
Wang, Y.-F., Y. Zhang, Z. Zhu, T.-Y. Wang, D. L. Morris, J. J. Shen, H. Zhang, H.-F. Pan, J. Yang, S. Yang, D.-Q. Ye, T. J. Vyse, Y. Cui, X. Zhang, Y. Sheng, Y. L. Lau, and W. Yang. 2018. Identification of ST3AGL4, MFHAS1, CSNK2A2 and CD226 as loci associated with systemic lupus erythematosus (SLE) and evaluation of SLE genetics in drug repositioning. Annals of the Rheumatic Diseases 77(7):1078–1084. http://dx.doi.org/10.1136/annrheumdis-2018-213093.
Wang, Y.-F., Y. Zhang, Z. Lin, H. Zhang, T.-Y. Wang, Y. Cao, D. L. Morris, Y. Sheng, X. Yin, S.-L. Zhong, X. Gu, Y. Lei, J. He, Q. Wu, J. J. Shen, J. Yang, T.-H. Lam, J.-H. Lin, Z.-M. Mai, M. Guo, Y. Tang, Y. Chen, Q. Song, B. Ban, C. C. Mok, Y. Cui, L. Lu, N. Shen, P. C. Sham, C. S. Lau, D. K. Smith, T. J. Vyse, X. Zhang, Y. L. Lau, and W. Yang. 2021. Identification of 38 novel loci for systemic lupus erythematosus and genetic heterogeneity between ancestral groups. Nature Communications 12(1):772. https://doi.org/10.1038/s41467-021-21049-y.
Wellcome Trust Case Control Consortium, and The Australo-Anglo-American Spondylitis Consortium (TASC). 2007. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nature Genetics 39(11):1329–1337. https://doi.org/10.1038/ng.2007.17.
Willer, C. J., D. A. Dyment, N. J. Risch, A. D. Sadovnick, G. C. Ebers, and G. Canadian Collaborative Study. 2003. Twin concordance and sibling recurrence rates in multiple sclerosis. Proceedings of the National Academy of Sciences of the United States of America 100(22):12877–12882. https://doi.org/10.1073/pnas.1932604100.
Wong, F. S., and L. Wen. 2003. The study of HLA class II and autoimmune diabetes. Current Molecular Medicine 3(1):1–15. https://doi.org/10.2174/1566524033361591.
Wu, X., L. Qian, K. Liu, J. Wu, and Z. Shan. 2021. Gastrointestinal microbiome and gluten in celiac disease. Annals of Medicine 53(1):1797–1805. https://doi.org/10.1080/07853890.2021.1990392.
Xie, F., H. Yun, S. Bernatsky, and J. R. Curtis. 2016. Brief report: Risk of gastrointestinal perforation among rheumatoid arthritis patients receiving tofacitinib, tocilizumab, or other biologic treatments. Arthritis & Rheumatology 68(11):2612–2617. https://doi.org/10.1002/art.39761.
Yannakakis, M. P., H. Tzoupis, E. Michailidou, E. Mantzourani, C. Simal, and T. Tselios. 2016. Molecular dynamics at the receptor level of immunodominant myelin oligodendrocyte glycoprotein 35-55 epitope implicated in multiple sclerosis. Journal of Molecular Graphics and Modelling 68:78–86. https://doi.org/10.1016/j.jmgm.2016.06.005.
Yofe, I., R. Dahan, and I. Amit. 2020. Single-cell genomic approaches for developing the next generation of immunotherapies. Nature Medicine 26(2):171–177. https://doi.org/10.1038/s41591-019-0736-4.
Young, K. A., M. E. Munroe, J. M. Guthridge, D. L. Kamen, T. B. Niewold, G. S. Gilkeson, M. H. Weisman, M. L. Ishimori, J. Kelly, P. M. Gaffney, K. H. Sivils, R. Lu, D. J. Wallace, D. R. Karp, J. B. Harley, J. A. James, and J. M. Norris. 2017. Combined role of vitamin D status and CYP24A1 in the transition to systemic lupus erythematosus. Annals of the Rheumatic Diseases 76(1):153–158. https://doi.org/10.1136/annrheumdis-2016-209157.
Zhang, F., K. Wei, K. Slowikowski, C. Y. Fonseka, D. A. Rao, S. Kelly, S. M. Goodman, D. Tabechian, L. B. Hughes, K. Salomon-Escoto, G. F. M. Watts, A. H. Jonsson, J. RangelMoreno, N. Meednu, C. Rozo, W. Apruzzese, T. M. Eisenhaure, D. J. Lieb, D. L. Boyle, A. M. Mandelin, 2nd, A. Accelerating Medicines Partnership Rheumatoid, C. Systemic Lupus Erythematosus, B. F. Boyce, E. DiCarlo, E. M. Gravallese, P. K. Gregersen, L. Moreland, G. S. Firestein, N. Hacohen, C. Nusbaum, J. A. Lederer, H. Perlman, C. Pitzalis, A. Filer, V. M. Holers, V. P. Bykerk, L. T. Donlin, J. H. Anolik, M. B. Brenner, and S. Raychaudhuri. 2019. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nature Immunology 20(7):928–942. https://doi.org/10.1038/s41590-019-0378-1.
Zhao, M., J. Jiang, M. Zhao, C. Chang, H. Wu, and Q. Lu. 2021. The application of single-cell rna sequencing in studies of autoimmune diseases: A comprehensive review. Clinical Reviews in Allergy & Immunology 60(1):68–86. https://doi.org/10.1007/s12016-020-08813-6.
Ziegler, A. G., P. Achenbach, R. Berner, K. Casteels, T. Danne, M. Gundert, J. Hasford, V. S. Hoffmann, O. Kordonouri, K. Lange, H. Elding Larsson, M. Lundgren, M. D. Snape, A. Szypowska, J. A. Todd, E. Bonifacio, and G. S. group. 2019. Oral insulin therapy for primary prevention of type 1 diabetes in infants with high genetic risk: The GPPAD-POInT (global platform for the prevention of autoimmune diabetes primary oral insulin trial) study protocol. BMJ Open 9(6):e028578. https://doi.org/10.1136/bmjopen-2018-028578.
Ziegler, A. G., M. Rewers, O. Simell, T. Simell, J. Lempainen, A. Steck, C. Winkler, J. Ilonen, R. Veijola, M. Knip, E. Bonifacio, and G. S. Eisenbarth. 2013. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA 309(23):2473–2479. https://doi.org/10.1001/jama.2013.6285.
Ziegler, S. M., C. N. Feldmann, S. H. Hagen, L. Richert, T. Barkhausen, J. Goletzke, V. Jazbutyte, G. Martrus, W. Salzberger, T. Renne, K. Hecher, A. Diemert, P. C. Arck, and M. Altfeld. 2018. Innate immune responses to Toll-like receptor stimulation are altered during the course of pregnancy. Journal of Reproductive Immunology 128:30–37. https://doi.org/10.1016/j.jri.2018.05.009.
This page intentionally left blank.


































































